CAC Antibody Collection
CAC Antibody Collection
The antibodies on this page are part of Cosmo Bio's exclusive CAC Collection. For many many thousands of other antibodies from many different makers, use our Search the Store function and our Explore Products drop down menu.
Antibodies for DNA Damage Research
| DNA Damage: UV-induced DNA lesions | |||
| Product name (click for order info) | Cat No (click for datasheet) |
Host | Species specificity |
| Anti Alpha-Crystallin B Chain, p19S pAb (Rabbit, Affinity Purified) | CAC-ACC-PA004 | RAB | HU MS RT BOV |
| Anti Alpha-Crystallin B Chain, p45S pAb (Rabbit, Affinity Purified) | CAC-ACC-PA005 | RAB | HU MS RT BOV |
| Anti Alpha-Crystallin B Chain, p59S pAb (Rabbit, Affinity Purified) | CAC-ACC-PA006 | RAB | HU RT BOV |
| DNA Damage: 8-Nitroguanosine for oxidative stress research | |||
| Product name (click for order info) | Cat No (click for datasheet) |
Host | Species specificity |
| Anti Keratin, Type I Cytoskeletal 18 (KRT18/Cytokeratin 18) mAb (Clone D2C7) | CAC-CE-046A | MS | HU MS RT MKY |
| Anti Tubulin Gamma-1 Chain (GCP-1) mAb (Clone E39) | CAC-NM-MA-003 | MS | HU MS RT |
| DNA Damage: Nucleotide excision repair factors | |||
| Product name (click for order info) | Cat No (click for datasheet) |
Host | Species specificity |
| Anti Cyclobutane Pyrimidine Dimers (CPDs) mAb (Clone TDM-2) | CAC-NM-DND-001 | MS | ALL |
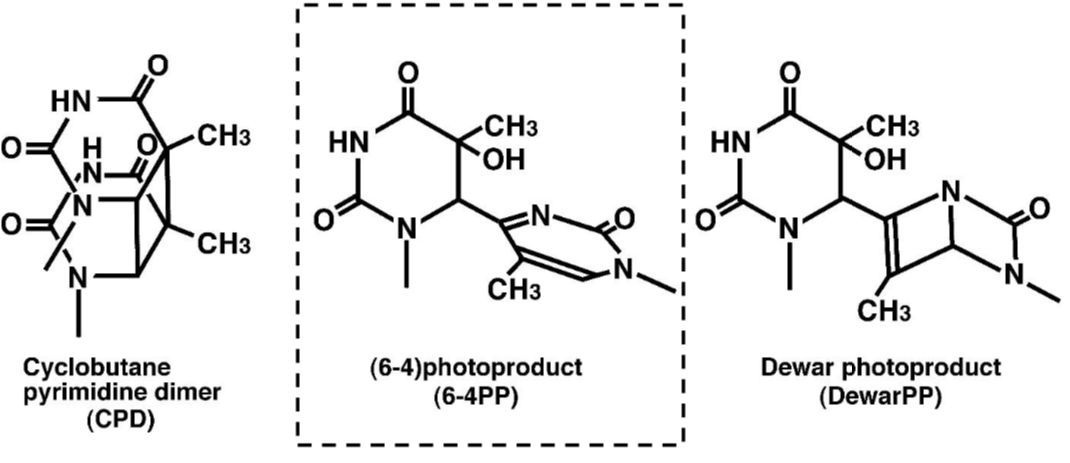
| Anti 6-4 Photoproducts (6-4PPs) mAb (Clone 64M-2) | CAC-NM-DND-002 | MS | ALL |
| Anti Dewar Photoproducts (DewarPPs) mAb (Clone DEM-1) | CAC-NM-DND-003 | MS | ALL |
| High Sensitivity 6-4PP/(6-4) Photoproduct ELISA Kit | CSR-NM-MA-K002 | MS | ALL |
| High Sensitivity CPD ELISA Kit Ver.2 | CSR-NM-MA-K003 | MS | ALL |
| Anti Acetylaminofluorene (AAF) DNA Adducts mAb (Clone AAF-1) | CAC-NM-MA-001 | MS | ALL |
|
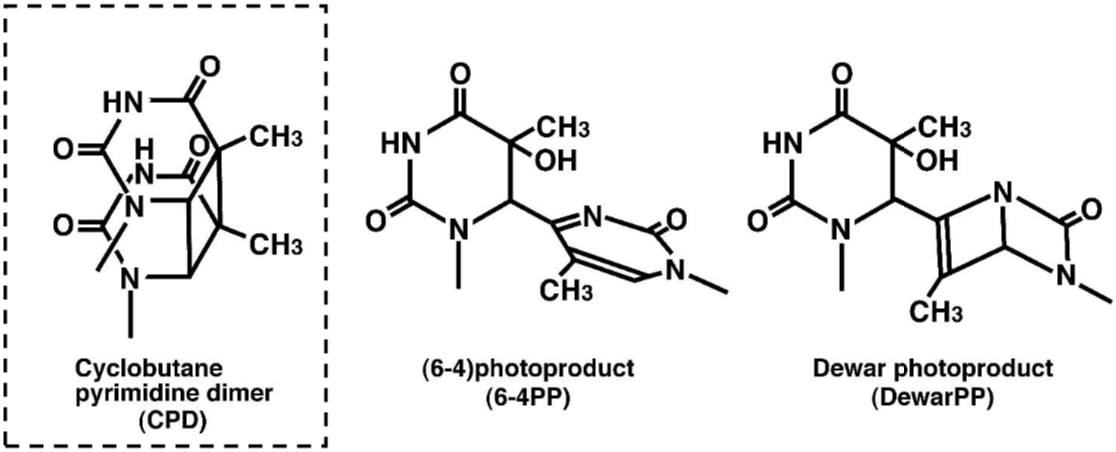
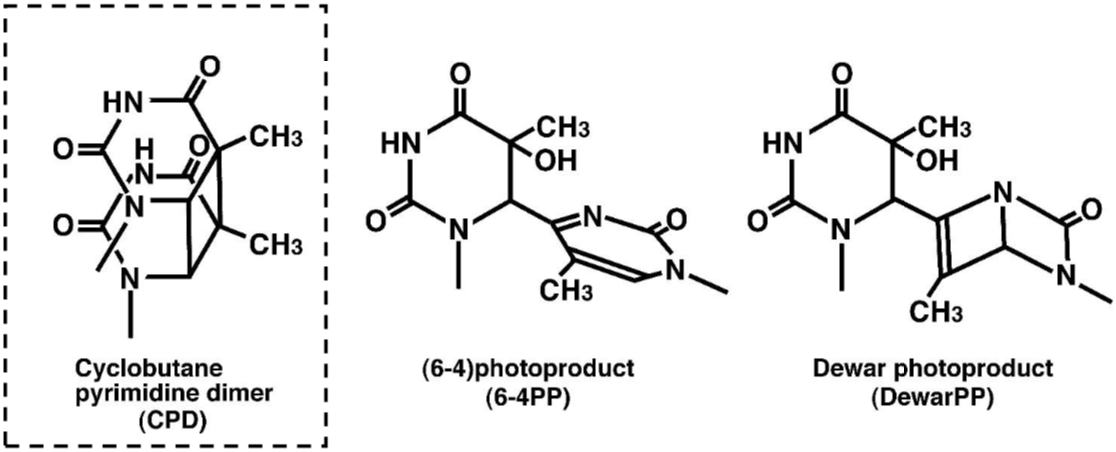
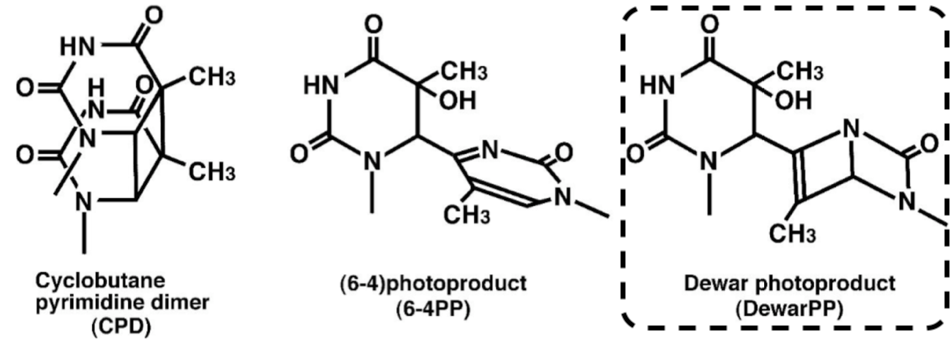
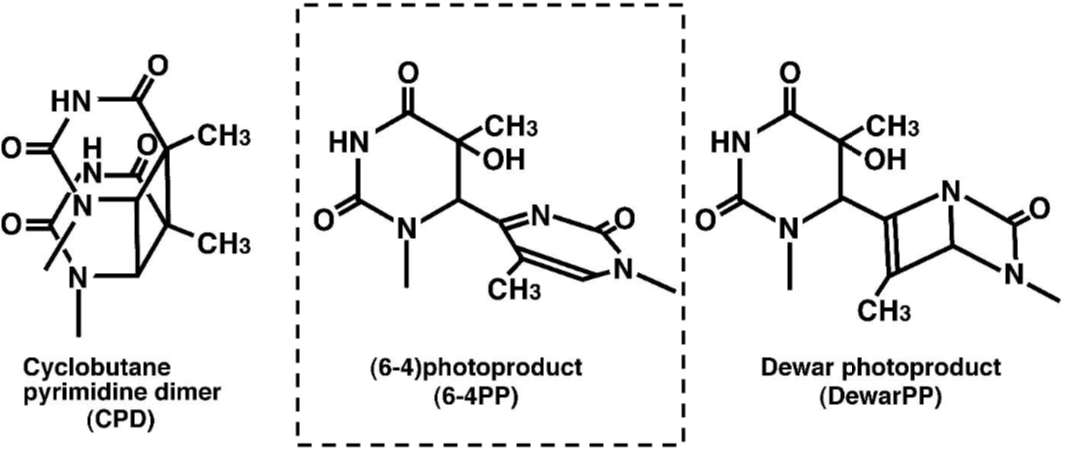
DNA damage in cells exposed to ultraviolet (UV) radiation plays significant roles in cell-cycle arrest, activation of DNA repair, cell killing, mutation, and neoplastic transformation. The major types of DNA damage induced by UVB (280-315 nm) and by UVC (200-280 nm) are cyclobutane pyrimidine dimers (CPDs) and (6-4) photoproducts (6-4PPs), which are formed between adjacent pyrimidine nucleotides on the same strand of DNA. Approximately 70-80% of UV-induced DNA damage is CPDs and the remainder are 6-4PPs and Dewar isomers of 6-4PPs. These types of DNA lesions are repaired by the nucleotide excision repair (NER) system in normal human cells. |
| Product name | Anti Cyclobutane Pyrimidine Dimers (CPDs) mAb (Clone TDM-2) |
| Cat No | CAC-NM-DND-001 (D) |
Description:
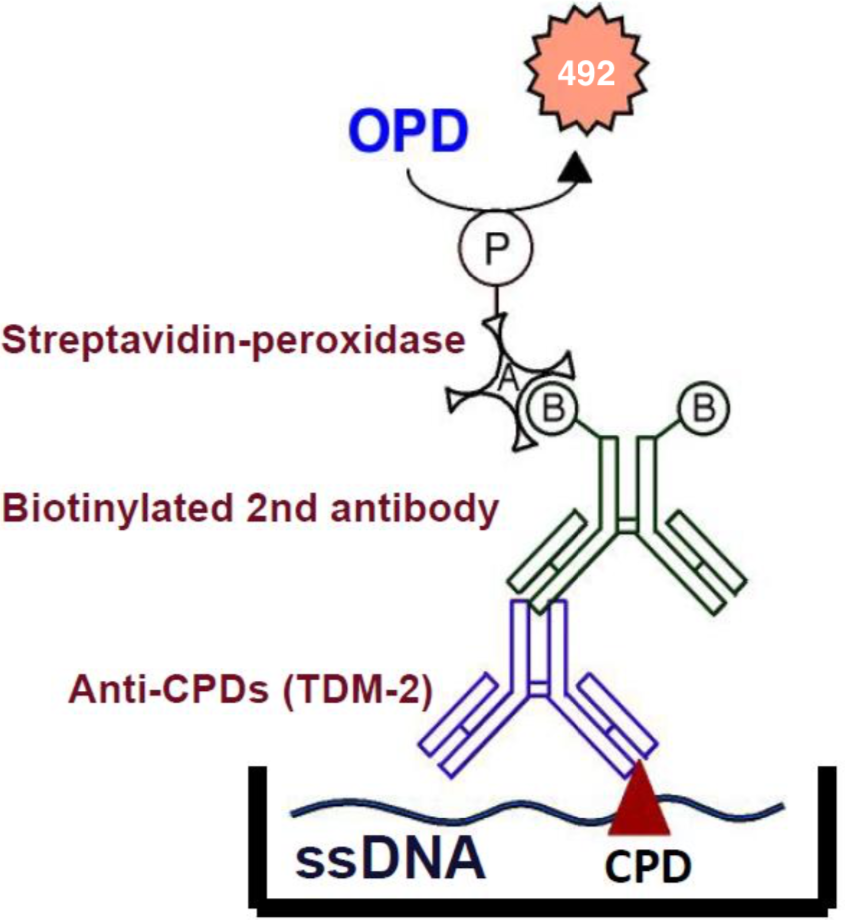
To better study molecular events surrounding UV-induced DNA damage and repair, Mori et al established monoclonal antibodies specific for CPDs or 6-4PPs. These antibodies enable one to quantitate photoproducts in DNA purified from cultured cells or from the skin epidermis using an enzyme-linked immunosorbent assay (ELISA). They can also be used in indirect immunofluorescence (IIF) to visualize and measure photoproducts in DNA of cultured cells or the skin. This technology will contribute to the understanding of molecular mechanisms of cellular responses to UV and DNA damage in many research fields including cancer, photobiology, dermatology, ophthalmology, immunology and cosmetology.
Reactivity:
1) CPDs in single-stranded DNA.
2) CPDs formed in every dipyrimidine sequence (TT, TC, CT and CC).
3) CPDs formed in oligonucleotides consisting of more than eight bases.
Host: Mouse
Species Specificity: All
References:
1) Yamamoto, A., et al., DNA Repair, 6, 649-657 (2007).
2) Matsumoto, M., et al., J. Cell Sci., 120, 1104-1112 (2007).
3) Yasuda, G., et al., Mol. Cell. Biol., 27, 6606-6614 (2007).
4) Sugasawa, K., et al., Cell 121, 387-400 (2005).
5) Nishiwaki, Y., et al., J. Invest. Dermatol. 122, 526-532 (2004).
6) Imoto, K., et al., J. Invest. Dermatol. 119, 1177-7782 (2002).
7) Wakasugi, M., et al., J. Biol. Chem., 277, 1637-1640 (2002).
8) Kobayashi, N., et al., Pigment Cell Res. 14, 94-102 (2001).
9) Katsumi, S., et al., J. Invest. Dermatol. 117, 1156-1161 (2001).
10) Otoshi, E., et al., Cancer Res. 60, 1729-1735 (2000).
11) Nakagawa, A., et al., J. Invest. Dermatol. 110, 143-148 (1998).
12) Kobayashi, N., et al., J. Invest. Dermatol. 110, 806-810 (1998).
13) Komatsu, Y., et al., Nucleic Acids Res. 25, 3889-3894 (1997).
14) Nakane, H., et al., Nature 377, 165-168 (1995).
15) Todo, T., et al., Nature 361, 371-374 (1993).
16) Kobayashi, N., et al., J. Invest. Dermatol. 101, 685-689 (1993).
17) Potten, C.S., et al., Int. J. Radiat. Biol. 63, 313-324 (1993).
18) Matsunaga, T., et al., Photochem. Photobiol. 54, 403-410 (1991).
19) Mori, T., et al., Photochem. Photobiol. 54, 225-232 (1991).
More than 200 papers using TDM-2 antibody have been published so far.
|
Figures: Figure 1: Structures of UV-induced DNA damage. |
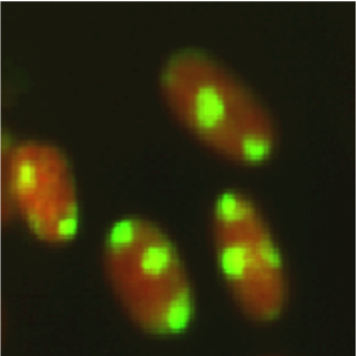
Figure 3: Fluorescent image of localized CPDs in normal human fibroblasts. Cells were cultured in a 35-mm glass-bottom dish for 24 hours. Immediately after micropore UV irradiation (100 J/m2), cells were fixed and permeabilized. After denaturation of DNA, CPDs (yellow) were visualized using immunofluorescence with clone TDM-2. Nuclear DNA (red) was counterstained with propidium iodide. A filter with 3-μm pores was used. |
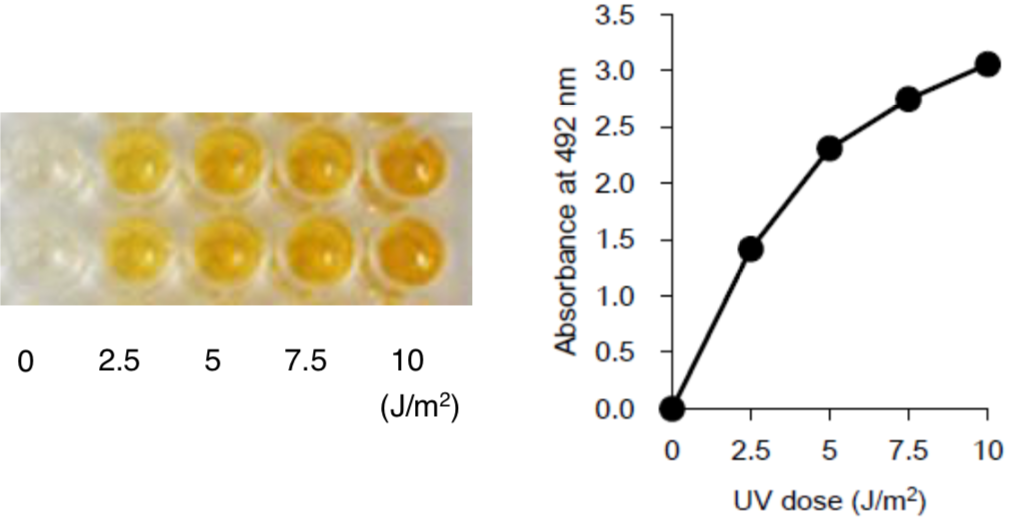
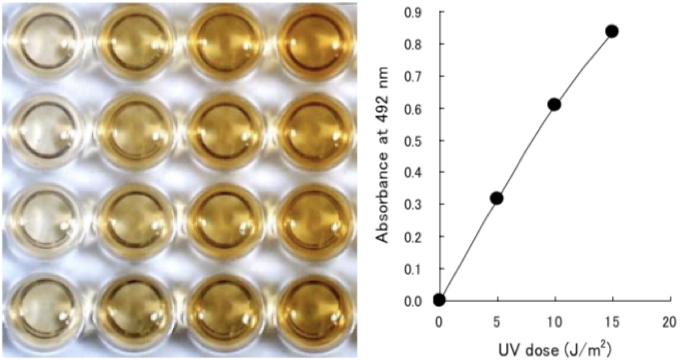
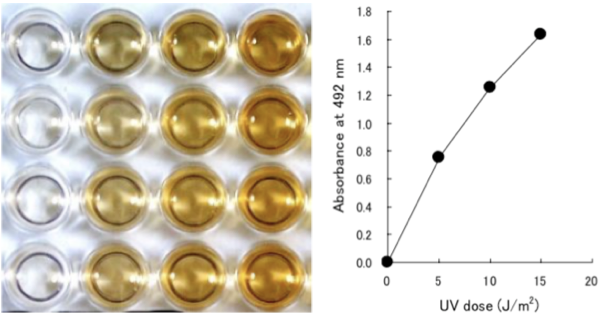 Figure 2: UV-induced CPDs are detected by ELISA. Dose-dependent induction of cyclobutane pyrimidine dimers (CPDs) in 254-nm UV-irradiated calf thymus DNA was measured by ELISA with clone TDM-2. |
| Product name | Anti 6-4 Photoproducts (6-4PPs) mAb (Clone 64M-2) |
| Cat No | CAC-NM-DND-002 |
|
Description REACTIVITY: |
|
References:
1) Yamamoto, A., et al., DNA Repair, 6, 649-657 (2007).
2) Matsumoto, M., et al., J. Cell Sci., 120, 1104-1112 (2007).
3) Yasuda, G., et al., Mol. Cell. Biol., 27, 6606-6614 (2007).
4) Sugasawa, K., et al., Cell 121, 387-400 (2005).
5) Nishiwaki, Y., et al., J. Invest. Dermatol. 122, 526-532 (2004).
6) Imoto, K., et al., J. Invest. Dermatol. 119, 1177-7782 (2002).
7) Wakasugi, M., et al., J. Biol. Chem., 277, 1637-1640 (2002).
8) Kobayashi, N., et al., Pigment Cell Res. 14, 94-102 (2001).
9) Katsumi, S., et al., J. Invest. Dermatol. 117, 1156-1161 (2001).
10) Otoshi, E., et al., Cancer Res. 60, 1729-1735 (2000).
11) Nakagawa, A., et al., J. Invest. Dermatol. 110, 143-148 (1998).
12) Kobayashi, N., et al., J. Invest. Dermatol. 110, 806-810 (1998).
13) Komatsu, Y., et al., Nucleic Acids Res. 25, 3889-3894 (1997).
14) Nakane, H., et al., Nature 377, 165-168 (1995).
15) Todo, T., et al., Nature 361, 371-374 (1993).
16) Kobayashi, N., et al., J. Invest. Dermatol. 101, 685-689 (1993).
17) Potten, C.S., et al., Int. J. Radiat. Biol. 63, 313-324 (1993).
18) Matsunaga, T., et al., Photochem. Photobiol. 54, 403-410 (1991).
19) Mori, T., et al., Photochem. Photobiol. 54, 225-232 (1991).
More than 200 papers using 64M-2 antibody have been published so far.
Host: Mouse
Species specificity: All
Figures
 |
 |
 |
| Figure 1 Structures of UV-induced DNA damage. |
Figure 2 UV-induced 6-4PPs are detected by ELISA. Dose-dependent induction of (6-4) photoproducts (6-4PPs) in 254-nm UV- irradiated calf thymus DNA was measured by ELISA with clone 64M-2. |
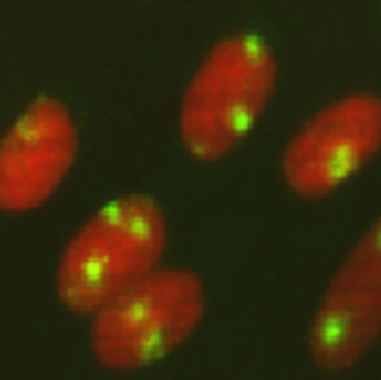
Figure 3 Fluorescent image of localized 6-4PPs in normal human fibroblasts. Cells were cultured in a 35-mm glass-bottom dish for 24 hours. Immediately after micropore UV irradiation (100 J/m2), cells were fixed and permeabilized. After denaturation of DNA, 6-4PPs (yellow) were visualized using immunofluorescence with clone 64M-2. Nuclear DNA (red) was counterstained with propidium iodide. A filter with 3-μm pores was used. |
| Product name | Anti Dewar Photoproducts (DewarPPs) mAb (Clone DEM-1) |
| Cat No | CAC-NM-DND-003 |
Description
To better study molecular events surrounding UV-induced DNA damage and repair, Matsunaga et al established monoclonal antibodies specific for DewarPPs. The antibodies enable one to quantitate photoproducts in DNA purified from cultured cells or from skin epidermis using an enzyme-linked immunosorbent assay (ELISA). They can also be used in indirect immunofluorescence (IIF) to visualize and measure photoproducts in DNA of cultured cells. This technology will contribute to understanding of molecular mechanisms of cellular responses to DewarPPs in many research fields including cancer, photobiology, dermatology, ophthalmology, immunology and cosmetology.
REACTIVITY:
1) DewarPPs in single-stranded DNA.
2) DewarPPs formed in TC, TT and CC dipyrimidine sequences.
3) DewarPPs formed in oligonucleotides consisting of more than eight bases.
References:
1) Douki, T. and Cadet. J, Biochemistry 40, 2495-2501 (2001).
2) Douki, T., et al., J. Biol. Chem., 275, 11678-11685 (2000).
3) Lee, J.H., et al., Proc. Natl. Acad. Sci. U.S.A., 97, 4591-4596 (2000).
4) Perdiz, D., et al., J. Biol. Chem. 275, 26732-26742 (2000).
5) Kobayashi, N., et al., J. Biochem. 123, 182-188 (1998).
6) Clingen, P.H., et al., Photochem. Photobiol. 61, 163-170 (1995).
7) Clingen, P.H., et al., Cancer Res. 55, 2245-2248 (1995).
8) Chadwick, C.A., et al., J. Photochem. Photobiol. B. 28, 163-170 (1995).
9) Matsunaga, T., et al., Photochem. Photobiol. 57, 934-940 (1993).
10) Matsunaga, T., et al., Photochem. Photobiol. 54, 403-410 (1991).
11) Mitchell D.L. Mutat. Res., 194, 227-237 (1988).
Host: Mouse
Species specificity: All
 |
 |
 |
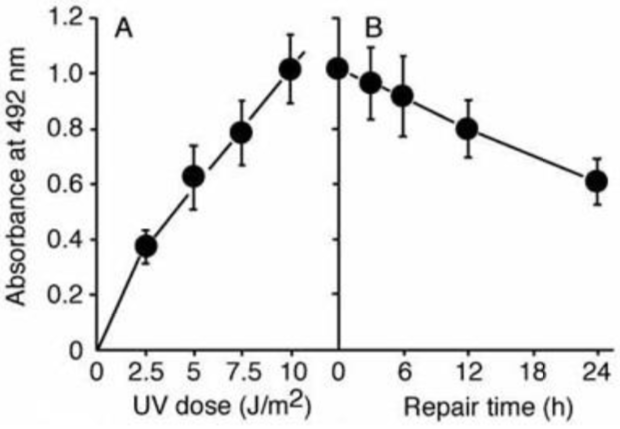
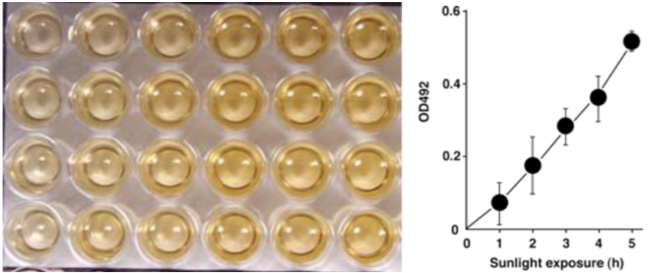
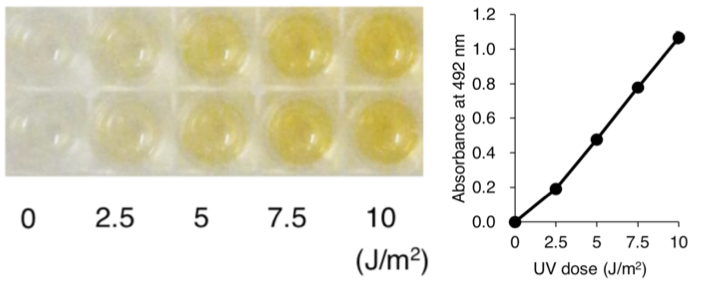
| Figure 1: Structures of UV-induced DNA damage. | Figure 2: Solar UV-induced DewarPPs are detected by ELISA. The exposure-dependent induction of Dewar photoproducts (DewarPPs) in solar UV-irradiated calf thymus DNA was measured by ELISA with clone DEM-1. |
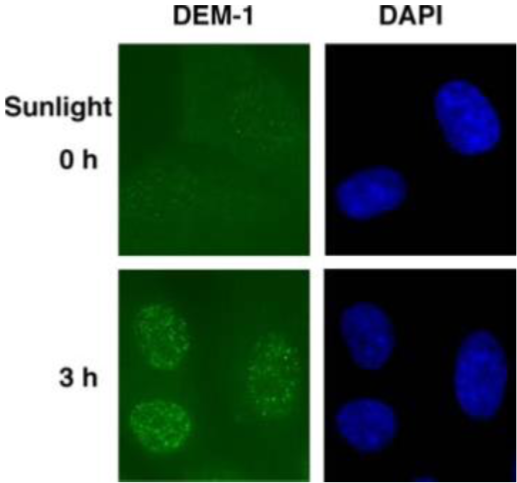
Figure 3: Fluorescent image of DewarPPs in normal human fibroblasts. Cells were cultured in a 35-mm glass-bottom dish for 24 hours. Immediately after solar UV irradiation for 3 hours or mock irradiation, cells were fixed and permeabilized. After denaturation of DNA, DewarPPs (yellow) were visualized using immunofluorescence with clone DEM-1. Nuclear DNA (blue) was counterstained with DAPI. |
| Product name | High Sensitivity 6-4 Photoproducts (6-4PPs) ELISA Kit |
| Cat No | CSR-NM-MA-K002 |
Description
To better study molecular events surrounding UV-induced DNA damage and repair, Mori et al. developed and characterized monoclonal antibodies (mAbs) specific for CPDs and for 6-4PPs while Matsunaga et al. developed and characterized mAbs specific for DewarPPs. Three of these antibodies (CPDs: clone TDM-2; 6-4PPs: clone 64M-2; DewarPPs: clone DEM-1) continue to be cited frequently in the literature, often for use in ELISA. This High Sensitivity (6-4) photoproducts (6-4PPs) ELISA Kit is the only commercially available ELISA utilizing anti-6-4PPs clone 64M-2 and has been optimized for highly sensitive detection of 6-4PPs in DNA purified from cultured cells or from skin epidermis. This ELISA detects 6-4PPs from dipyrimidines in all DNA sequence contexts (i.e., TT, TC, CT and CC). Thus, the availability and convenience of this ELISA Kit will contribute to further understanding of molecular mechanisms involved in cellular responses to UV radiation and DNA damage with applications across many research fields including cancer, photobiology, dermatology, ophthalmology, immunology and cosmetics science.
References:
1) Yamamoto, A., et al., DNA Repair, 6, 649-657 (2007).
2) Matsumoto, M., et al., J. Cell Sci., 120, 1104-1112 (2007).
3) Yasuda, G., et al., Mol. Cell. Biol., 27, 6606-6614 (2007).
4) Sugasawa, K., et al., Cell 121, 387-400 (2005).
5) Nishiwaki, Y., et al., J. Invest. Dermatol. 122, 526-532 (2004).
6) Imoto, K., et al., J. Invest. Dermatol. 119, 1177-7782 (2002).
7) Wakasugi, M., et al., J. Biol. Chem., 277, 1637-1640 (2002).
8) Kobayashi, N., et al., Pigment Cell Res. 14, 94-102 (2001).
9) Katsumi, S., et al., J. Invest. Dermatol. 117, 1156-1161 (2001).
10) Otoshi, E., et al., Cancer Res. 60, 1729-1735 (2000).
11) Nakagawa, A., et al., J. Invest. Dermatol. 110, 143-148 (1998).
12) Kobayashi, N., et al., J. Invest. Dermatol. 110, 806-810 (1998).
13) Komatsu, Y., et al., Nucleic Acids Res. 25, 3889-3894 (1997).
14) Nakane, H., et al., Nature 377, 165-168 (1995).
15) Todo, T., et al., Nature 361, 371-374 (1993).
16) Kobayashi, N., et al., J. Invest. Dermatol. 101, 685-689 (1993).
17) Potten, C.S., et al., Int. J. Radiat. Biol. 63, 313-324 (1993).
18) Matsunaga, T., et al., Photochem. Photobiol. 54, 403-410 (1991).
19) Mori, T., et al., Photochem. Photobiol. 54, 225-232 (1991).
More than 200 papers using TDM-2 antibody have been published so far.
Host: Mouse
Species specificity: All
 |
 |
 |
 |
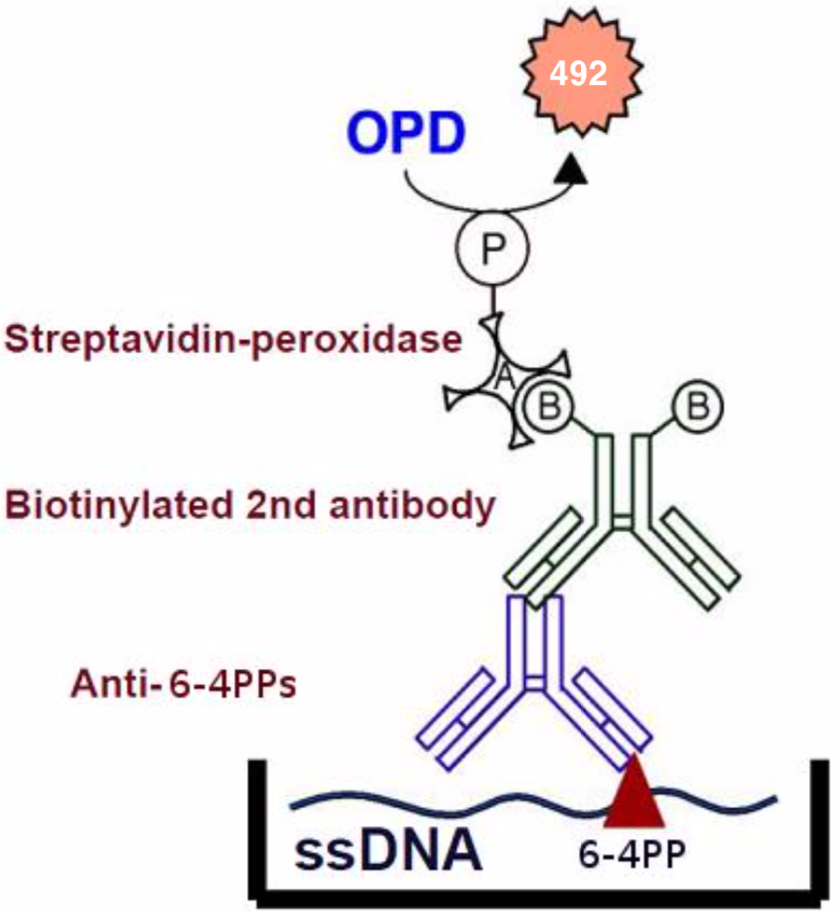
| Figure1:Structures of UV-induced DNA damage. | Figure 2: An ELISA for 6-4PPs. 1) Anti-6-4PPs monoclonal antibody clone 64M-2 recognizes 6-4PPs on single-stranded DNA. 2) 64M-2 binds to 6-4PPs formed each dipyrimidine sequence context (TT, TC, CT and CC). 3) 64M-2 stably binds to 6-4PPs in DNA longer than eight bases. 4) 64M-2 binds to 6-4PPs in UV-irradiated DNA purified from a wide range of sources. |
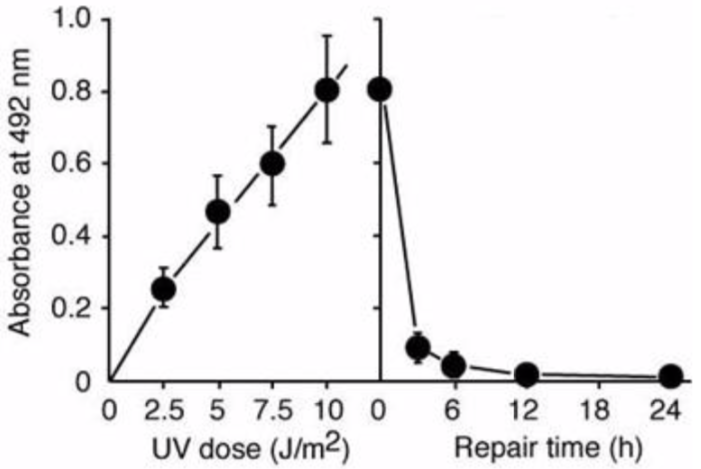
Figure 3: UV-induced 6-4PPs in DNA measured by ELISA. | Figure 4: Formation and repair of UV-induced 6-4PPs in human cells measured by ELISA. UVC radiation induces 6-4PPs HeLa cell DNA in a dose-dependent manner. The initial level of 6-4PPs induced by 10 J/m2 of UVC decreases over time as 6-4PPs are repaired, indicating the capacity of nucleotide excision repair in HeLa cells. |
| Product name | High Sensitivity Cyclobutane Pyrimidine Dimers (CPDs) ELISA Kit Ver.2 |
| Cat No | CSR-NM-MA-K003 |
Description
To better study molecular events surrounding UV-induced DNA damage and repair, Mori et al. developed and characterized monoclonal antibodies (mAbs) specific for CPDs and for 6-4PPs while Matsunaga et al. developed and characterized mAbs specific for DewarPPs. Three of these antibodies (CPDs: clone TDM-2; 6-4PPs: clone 64M-2; DewarPPs: clone DEM-1) continue to be cited frequently in the literature, often for use in ELISA. This High Sensitivity Cyclobutane Pyrimidine Dimers (CPDs) ELISA Kit is the only commercially available ELISA utilizing anti-CPDs clone TDM-2 and has been optimized for highly sensitive detection of CPDs in DNA purified from cultured cells or from skin epidermis. This ELISA detects CPDs from dipyrimidines in all DNA sequence contexts (i.e., TT, TC, CT and CC). Thus, the availability and convenience of this ELISA Kit will contribute to further understanding molecular mechanisms involved in cellular responses to UV radiation and DNA damage with applications across many research fields including cancer, photobiology, dermatology, ophthalmology, immunology and cosmetics science.
References:
1) Yamamoto, A., et al., DNA Repair, 6, 649-657 (2007).
2) Matsumoto, M., et al., J. Cell Sci., 120, 1104-1112 (2007).
3) Yasuda, G., et al., Mol. Cell. Biol., 27, 6606-6614 (2007).
4) Sugasawa, K., et al., Cell 121, 387-400 (2005).
5) Nishiwaki, Y., et al., J. Invest. Dermatol. 122, 526-532 (2004).
6) Imoto, K., et al., J. Invest. Dermatol. 119, 1177-7782 (2002).
7) Wakasugi, M., et al., J. Biol. Chem., 277, 1637-1640 (2002).
8) Kobayashi, N., et al., Pigment Cell Res. 14, 94-102 (2001).
9) Katsumi, S., et al., J. Invest. Dermatol. 117, 1156-1161 (2001).
10) Otoshi, E., et al., Cancer Res. 60, 1729-1735 (2000).
11) Nakagawa, A., et al., J. Invest. Dermatol. 110, 143-148 (1998).
12) Kobayashi, N., et al., J. Invest. Dermatol. 110, 806-810 (1998).
13) Komatsu, Y., et al., Nucleic Acids Res. 25, 3889-3894 (1997).
14) Nakane, H., et al., Nature 377, 165-168 (1995).
15) Todo, T., et al., Nature 361, 371-374 (1993).
16) Kobayashi, N., et al., J. Invest. Dermatol. 101, 685-689 (1993).
17) Potten, C.S., et al., Int. J. Radiat. Biol. 63, 313-324 (1993).
18) Matsunaga, T., et al., Photochem. Photobiol. 54, 403-410 (1991).
19) Mori, T., et al., Photochem. Photobiol. 54, 225-232 (1991).
More than 200 papers using TDM-2 antibody have been published so far.
Host: Mouse
Species specificity: All
Figures
|
Figure1:Structures of UV-induced DNA damage. |
Figure 2: An ELISA for CPDs. |
|
|
Figure 3: UV-induced CPDs in DNA measured by ELISA. |
||
|
Figure 4: Formation and repair of UV-induced CPDs in human cells measured by ELISA. |
||
| Product name | Anti Acetylaminofluorene (AAF) DNA Adducts mAb (Clone AAF-1) |
| Cat No | CAC-NM-MA-001 |
Description
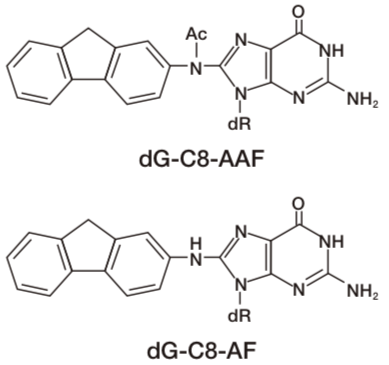
DNA adducts in mammalian cells exposed to N-acetoxy-2-acetylaminofluorene (NA-AAF), an activated derivative of the potent carcinogen 2-AAF, play significant roles in cell killing, chromosomal aberrations, gene mutations and neoplastic transformation. NA-AAF binds covalently to guanine in the DNA of mammalian cells and produces three different DNA adducts. The C-8 adducts dG-C8-AAF and deacetylated dG-C8-AF account for the majority of the DNA-bound products, while the minor N2 adduct dG-N2-AAF accounts for the remainder. The relative induction levels of the two major C-8 adducts vary among cell types. These adducts distort the DNA helix and therefore are repaired by nucleotide excision repair in human cells. Our AAF-1 antibodies bind most efficiently to dG-C8-AAF and less efficiently to dG-C8-AF in denatured DNA. The antibodies enable one to detect AAF-DNA adducts in DNA from cultured cells using an enzyme-linked immunosorbent assay (ELISA) and to visualize them in cultured cells or rodent tissues by immunofluorescence (IF). This technology will contribute to understanding of molecular mechanisms in AAF-related research fields including cancer and toxicology. Source: Toshio Mori Professor, Research Institute for Advanced Medicine, Nara Medical University.
References:
1) R.H. Heflich and R.E. Neft, Genetic toxicity of 2-acetylaminofluorene, 2-aminofluorene and some of their metabolites and model metabolites. Mutation Res. 318 (1994) 73-174.
2) E.Kriek, Fifty years of research on N-acetyl-2-aminofluorene, one of the most versatile compounds in experimental cancer research. J. Cancer Res. Clin. Oncol. 118 (1992) 481-489.
3) T. Iwamoto et al., In situ detection of acetylaminofluorene-DNA adducts in human cells using monoclonal antibodies. DNA Repair 3 (2004) 1475-1482.
Host: Mouse
Species specificity: All
Figures
 |
 |
 |
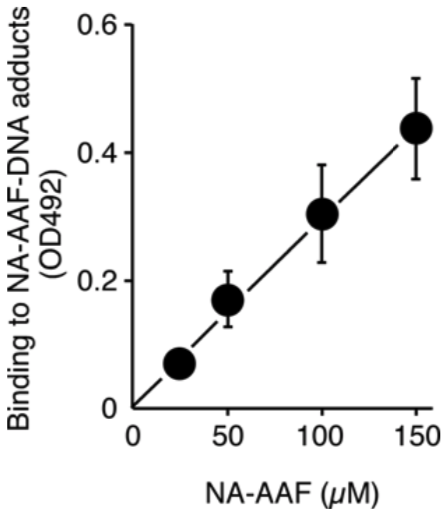
| Figure1: AAF-DNA adducts recognized by AAF-1. | Figure 2: Dose-dependent formation of NA-AAF-induced DNA adducts in human cells. Cells were exposed to NA-AAF for 0.5 h and the formation of DNA adducts in denatured DNA (500 ng/well) was determined using a sensitive-direct-binding ELISA with AAF-1 (1/100). (Details are shown in Ref. 3) |
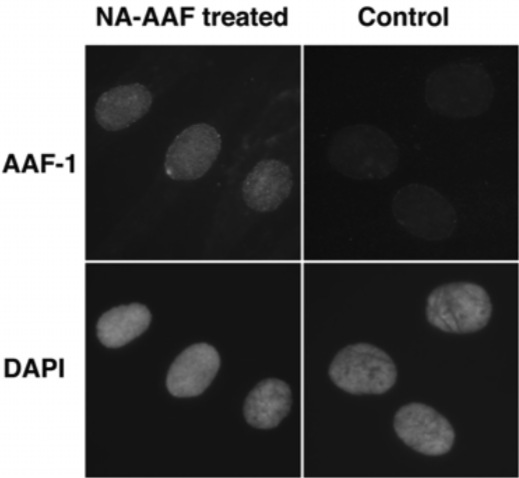
Figure 3: The formation of NA-AAF-induced DNA adducts in human cells. Cells were exposed to 200 μM NA-AAF or solvent for 0.5 h. After permeabilization and fixation, DNA adducts were visualized by sequential treatment with AAF-1 (1/25) and ALEXA FLUOR® 488 goat anti-mouse IgG conjugate. Nuclear DNA was counterstained with DAPI. |
| Product name | Anti 8-Nitroguanosine mAb (Clone NO2G52) |
| Cat No | CAC-KMU-M01 |
Description
Anti-Nitroguanosine monoclonal antibody (NO2G52), does not cross-react with normal nucleotide bases but selectively reacts with nitro functionalized nucleotides such as nitro-cGMP, nitro-GMP, and nitro-GTP. NO2G52 is commonly used for immunostaining. In addition, NO2G52 can be utilized for affinity purification of nitroguanine derivatives.
References:
1) T. Akaike, S. Okamoto, T. Sawa, J. Yoshitake, F. Tamura, K. Ichimori, K. Miyazaki, K. Sasamoto and H. Maeda, 8-nitroguanosine formation in viral pneumonia and its implication for pathogenesis, Proc. Natl. Acad. Sci. USA, 100, 685-690 (2003).
2) J. Yoshitake, T. Akaike, T. Akuta, F. Tamura, T. Ogura, H. Esumi, and H. Maeda, Nitric oxide as an endogenous mutagen for Sendai virus without antiviral activity, J. Virol., 78, 8709-8719 (2004).
3) T. Sawa, M. H. Zaki, T. Okamoto, T. Akuta, Y. Tokutomi, S. Kim-Mitsuyama, H. Ihara, A. Kobayashi, M. Yamamoto, S. Fujii, H. Arimoto, and T. Akaike, Protein S-guanylation by the biological signal 8-nitroguanosine 3',5'-cyclic monophosphate, Nat. Chem. Biol., 3, 727-735 (2007).
4) M. H.Zaki, S. Fujii, T. Okamoto, S. Islam, S. Khan, K. A. Ahmed, T. Sawa, and T. Akaike, Cytoprotective function of heme oxygenase 1 induced by a nitrated cyclic nucleotide formed during murine salmonellosis, J. Immunol., 182, 3746-3756 (2009).
5) Y. Terasaki, T. Akuta, M. Terasaki, T. Sawa, T. Mori, T. Okamoto, M. Ozaki, M. Takeya and T. Akaike, Guanine nitration in idiopathic pulmonary fibrosis and its implication for carcinogenesis, Am. J. Respir. Crit. Care. Med., 174, 665-673 (2006).
6) T. Sawa, M. Tatemichi, T. Akaike, A. Barbin and H. Ohshima, Analysis of urinary 8-nitroguanine, a marker of nitrative nucleic acid damage, by high-performance liquid chromatography-electrochemical detection coupled with immunoaffinity purification: association with cigarette smoking, Free Radic. Biol. Med., 40, 711-720 (2006).
7) M. Feelisch, Nitrated cyclic GMP as a new cellular signal, Nat. Chem. Biol., 3, 687-688 (2007).
8) K. A. Ahmed, T. Sawa, T. Akaike, Protein cysteine S-guanylation and electrophilic signal transduction by endogeneous nitro-nucleotides, Amino Acids, (2010).
9) T. Sawa, H. Arimoto and T. Akaike, Regulation of redox signaling involving chemical conjugation of protein thiols by nitric oxide and electrophiles, Bioconjugate Chem. (2010).
Host Mouse
Species specificity All
Figures
 |
 |
 |
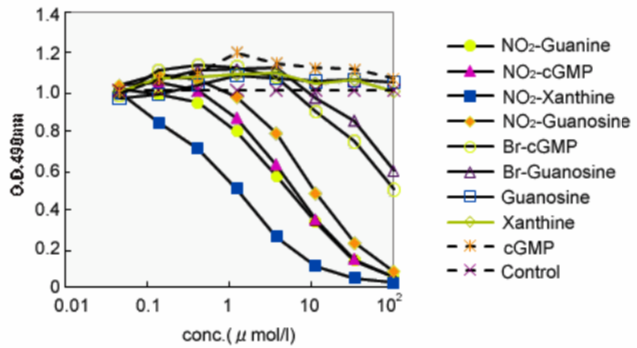
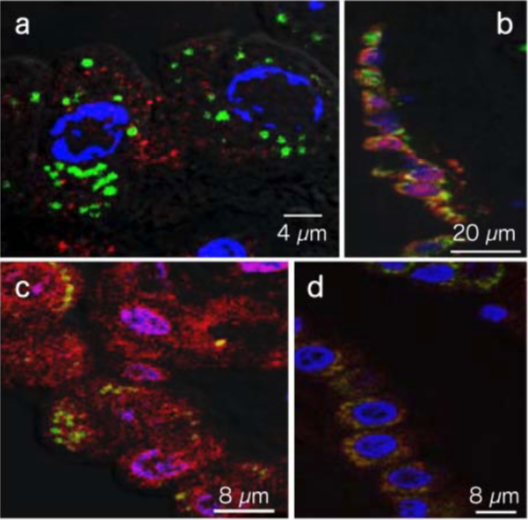
| Figure1:ELISA analysis of target specificity of monoclonal antibody NO2G52. | Figure 2: Immunofluorescence analysis of 8-nitroguanine production by airway epithelial cells from idiopathic Fibrosis (IPS) patients. A. 8-nitroguanine (green) and iNOS (red) B. Bright-field C. 8-nitroguanine (green) and 8-oxoguanine (red) D. 8-nitroguanine (green) and mitochondria (red) |
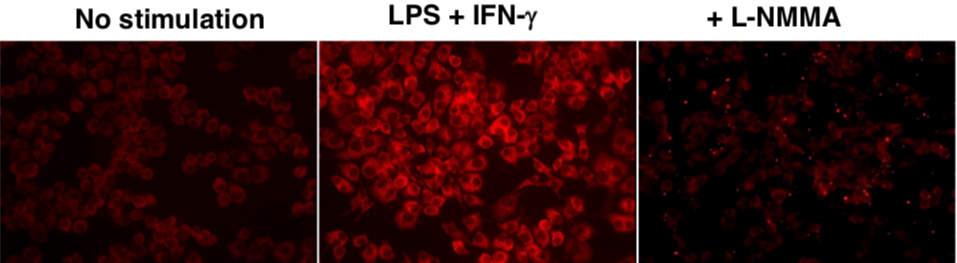
Figure 3: Immunofluorescence analysis of endogenous guanine nitration in murine macrophage cell line RAW 264.7 |
| Product name | Anti 8-Nitroguanosine pAb (Rabbit) |
| Cat No | CAC-KMU-P01 |
Description
Anti-Nitroguanosine polyclonal antibody does not cross-react with normal nucleobases but selectively reacts with nitrated nucleic acids such as nitroguanosine, nitroguanine, and nitroxanthine. Therefore, Anti-Nitroguanosine polyclonal antibody is a universal antibody against guanine modified 8th position with a nitro group. Anti-Nitroguanosine polyclonal antibody has very high affinity for 8-nitroguanine and 8-nitroguanosine, but it does not cross-react with normal guanosine, guanine, 8-hydroxyguanine or 3-nitrotyrosine. Since this antibody was prepared using rabbits, it can be used for immuno-histostaining of rodent tissues.
References:
1) T. Akaike, S. Okamoto, T. Sawa, J. Yoshitake, F. Tamura, K. Ichimori, K. Miyazaki, K. Sasamoto and H. Maeda, 8-nitroguanosine formation in viral pneumonia and its implication for pathogenesis, Proc. Natl. Acad. Sci. USA, 100, 685-690 (2003).
2) J. Yoshitake, T. Akaike, T. Akuta, F. Tamura, T. Ogura, H. Esumi, and H. Maeda, Nitric oxide as an endogenous mutagen for Sendai virus without antiviral activity, J. Virol., 78, 8709-8719 (2004).
3) T. Sawa, M. H. Zaki, T. Okamoto, T. Akuta, Y. Tokutomi, S. Kim-Mitsuyama, H. Ihara, A. Kobayashi, M. Yamamoto, S. Fujii, H. Arimoto, and T. Akaike, Protein S-guanylation by the biological signal 8-nitroguanosine 3',5'-cyclic monophosphate, Nat. Chem. Biol., 3, 727-735 (2007).
4) M. H.Zaki, S. Fujii, T. Okamoto, S. Islam, S. Khan, K. A. Ahmed, T. Sawa, and T. Akaike, Cytoprotective function of heme oxygenase 1 induced by a nitrated cyclic nucleotide formed during murine salmonellosis, J. Immunol., 182, 3746-3756 (2009).
5) Y. Terasaki, T. Akuta, M. Terasaki, T. Sawa, T. Mori, T. Okamoto, M. Ozaki, M. Takeya and T. Akaike, Guanine nitration in idiopathic pulmonary fibrosis and its implication for carcinogenesis, Am. J. Respir. Crit. Care. Med., 174, 665-673 (2006).
6) T. Sawa, M. Tatemichi, T. Akaike, A. Barbin and H. Ohshima, Analysis of urinary 8-nitroguanine, a marker of nitrative nucleic acid damage, by high-performance liquid chromatography-electrochemical detection coupled with immunoaffinity purification: association with cigarette smoking, Free Radic. Biol. Med., 40, 711-720 (2006).
7) M. Feelisch, Nitrated cyclic GMP as a new cellular signal, Nat. Chem. Biol., 3, 687-688 (2007).
8) K. A. Ahmed, T. Sawa, T. Akaike, Protein cysteine S-guanylation and electrophilic signal transduction by endogeneous nitro-nucleotides, Amino Acids (2010).
9) T. Sawa, H. Arimoto and T. Akaike, Regulation of redox signaling involving chemical conjugation of protein thiols by nitric oxide and electrophiles, Bioconjugate Chem (2010).
Host Rabbit
Species specificity All
Figures
 |
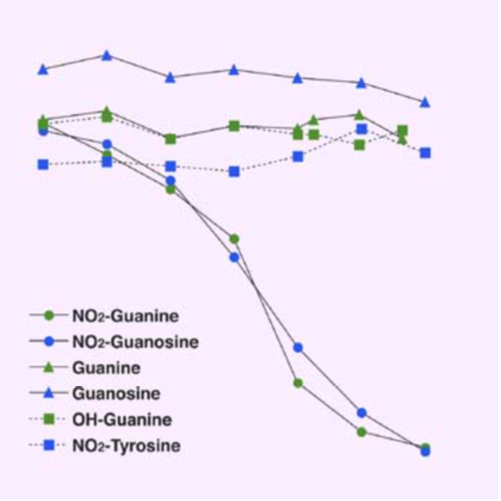 |
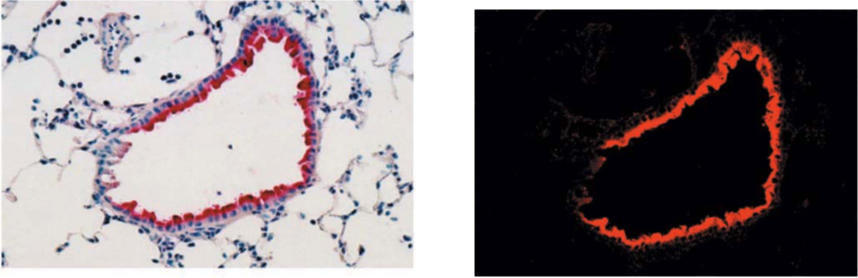
| Figure1:Immunostaining of influenza virus-infected mouse lung epithelial cells | Figure 2: ELISA analysis of anti-Nitroguanosine polyclonal antibody specificity |
| Product name | Anti DNA Repair Protein Complementing XP-A Cells (XPA) mAb (Clone A-2) |
| Cat No | CAC-KUP-TM-M01 |
Description:
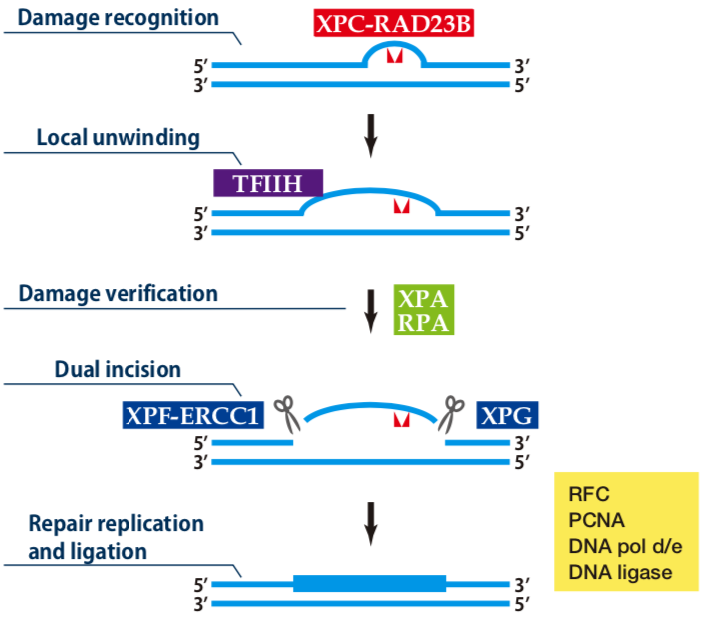
XPA has an ability to bind to DNA with some preference to damaged DNA and interacts with most of other NER factors. XPA appears to be involved in a proper assembly of preincision complex and verification of damaged DNA strand.
References:
Nagao A, Zhao X, Takegami T, et al., Multiple shRNA expressions in a single plasmid vector improve RNAi against the XPA gene. Biochem. Biophys. Res. Commun. 370, 301-305, 2008.
Host Mouse
Species specificity HU
Figures
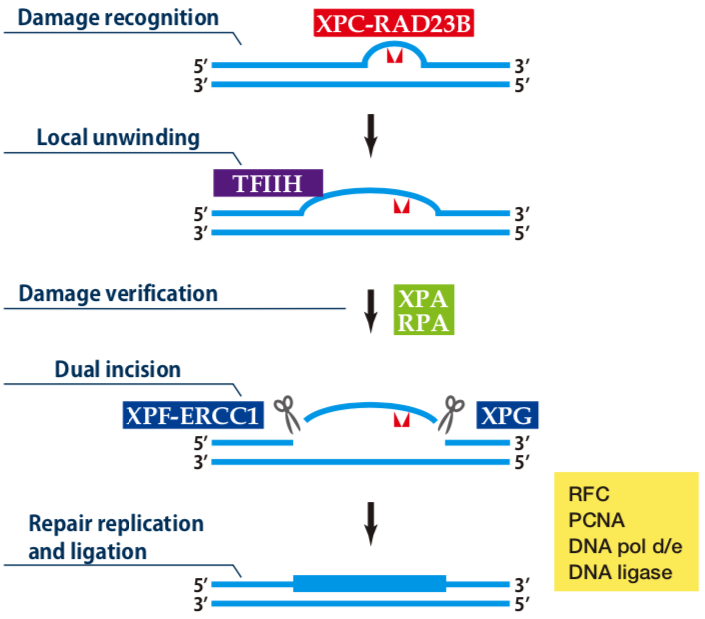 |
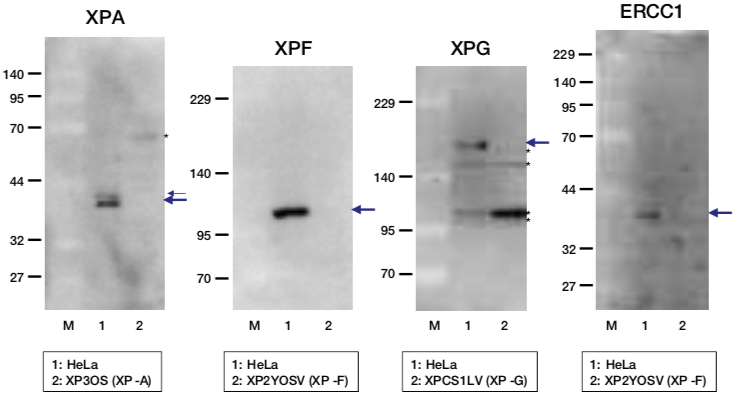 |
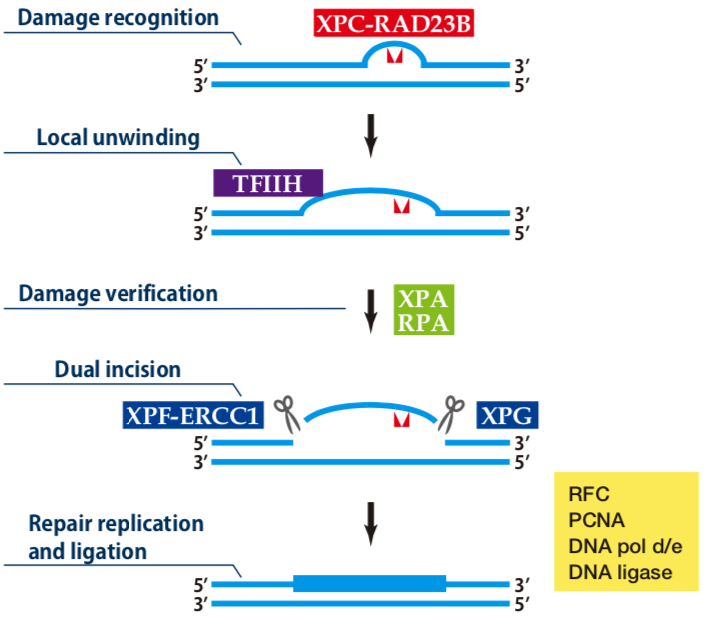
| Figure1: Current Model for the Dual Incision Process of NER | Figure 2: Western blot detection using nucleotide excision repair factor related antibodies |
| Product name | Anti DNA Repair Protein Complementing XP-A Cells (XPA) mAb (Clone 5F12) |
| Cat No | BAM-70-031-EX |
Description
XP (Xeroderma pigmentosum) is an autosomal recessive human disease characterized by hypersensitivity to sunlight and a high incidence of skin cancer on sun-exposed skin Cells from XP patients are hypersensitive to killing by UV irradiation because of a defect in nucleotide excision repair (NER). XP is classified into seven complementation groups (A-G) and a variant form. XPA shows the most severe symptoms. Products encoded by the XP genes function in repairing UV-induced cyclobutane pyrimidine dimmer and (6-4) photoproducts as well as chemically induced variety of DNA lesions. XPA protein consists of 273 amino acids and forms a complex with many proteins such as RPA, ERCC1, TFIIH, XAB1, and XAB2, which play roles in early steps of NER. The hybridoma 5F12 was constructed by Prof. K. Tanaka’s group who first cloned the XPA gene.
References:
1) Friedberg E C et. al., DNA Repair and Mutagenesis 2nd ed., ASM Press (2006).
2) Saijo M et al “Inhibition of nucleotide excision repair by anti-XPA monoclonal antibodies which interfere with
binding to RPA, ERCC1, and TFIIH” Biochem Biophys Res Comm 321:815-822 (2004). PMID: 15358100
3) Tanaka K et al “Analysis of a human DNA excision repair gene involved in group A xeroderma pigmentosum
and containing a zinc-finger domain” Nature 348:73 -76 (1990). PMID: 2234061
Host Mouse
Species specificity HU
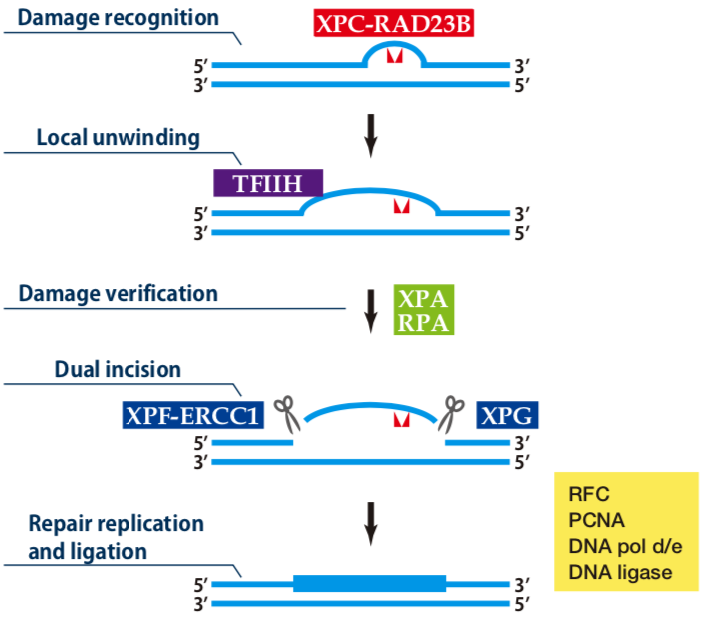 |
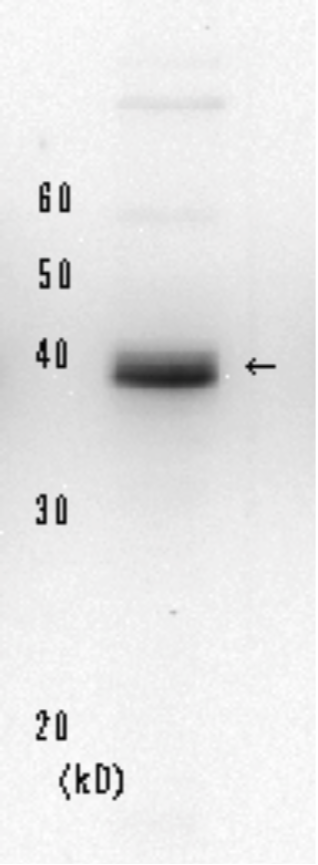 |
| Figure1: Current Model for the Dual Incision Process of NER | Figure 2: Detection of XPA protein in the crude extract of HeLa cells by Western blotting using clone 5F12 |
| Product name | Anti DNA Repair Endonuclease XPF (XPF/ERCC4) mAb (Clone 19-16) |
| Cat No | CAC-KUP-TM-M02 |
Description XPF harbors a nuclease domain and forms a stable complex with ERCC1. The ERCC1-XPF complex has a unique ability to make a nick on the DNA strand which makes the transition from duplex to single-stranded DNA in the 5' to 3' direction. In the NER process, ERCC1-XPF is responsible for 5'-incision at a dual incision step.
Host: Mouse
Species specificity: HU MS
 |
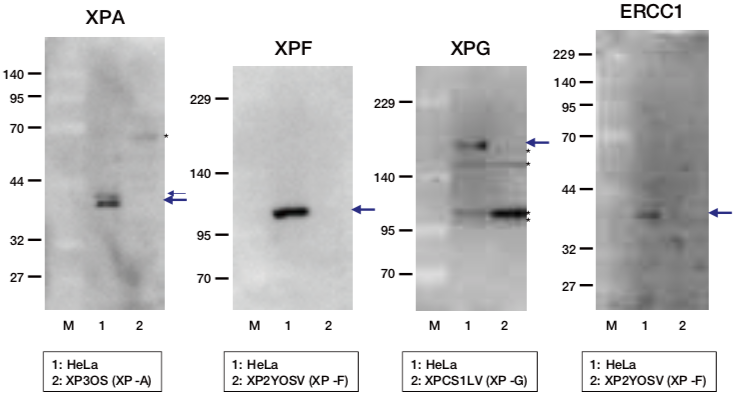 |
| Figure1: Current Model for the Dual Incision Process of NER. | Figure 2: Western blot detection using nucleotide excision repair factor related antibodies. |
| Product name | Anti DNA Repair Protein Complementing XP-G Cells (XPG/ERCC5) mAb (Clone G-26) |
| Cat No | CAC-KUP-TM-M03 |
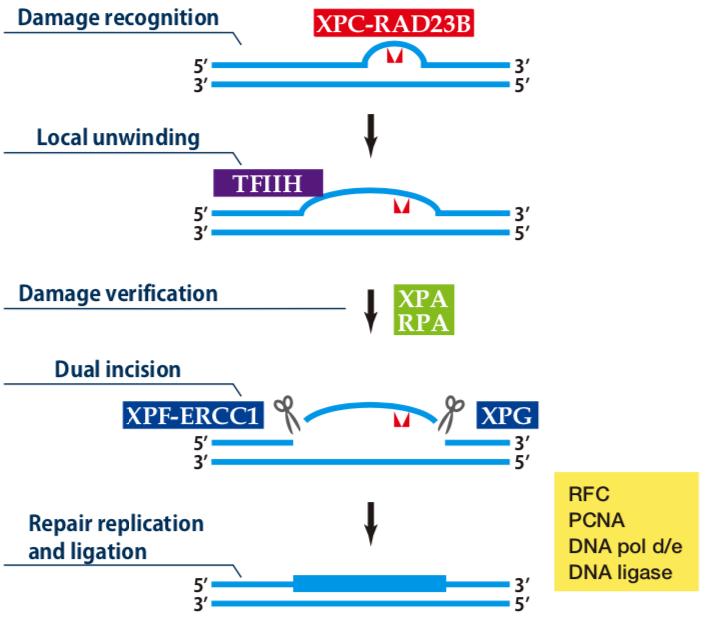
Description XPG is a structure-specific endonuclease with an opposite polarity to ERCC1-XPF and makes a nick on the DNA strand which makes the transition from single-stranded to duplex DNA in the 5' to 3' direction. In the NER process, XPG is responsible for 3'-incision at a dual incision step.
References:
Oyama M, Wakasugi M, Hama T, et al., Human NTH1 physically interacts with p53 and proliferating cell nuclear antigen. Biochem. Biophys. Res. Commun. 321, 183-191 (2004).
Host Mouse
Species specificity HU
 |
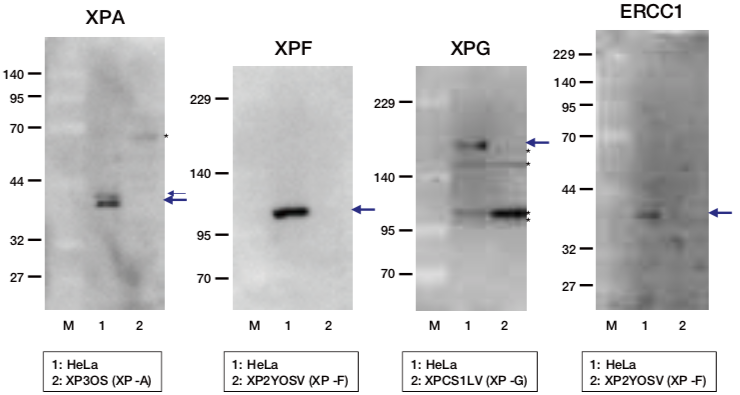 |
| Figure1:Current Model for the Dual Incision Process of NER. | Figure 2: Western blot detection using nucleotide excision repair factor related antibodies. |
| Product name | Anti DNA Excision Repair Protein ERCC-1 (ERCC1) mAb (Clone E1-44) |
| Cat No | CAC-KUP-TM-M04 |
Description:
ERCC1 forms a stable complex with XPF and the heterodimer has an ability to make a nick on the DNA strand which makes the transition from duplex to single-stranded DNA in the 5' to 3' direction. In the NER process, ERCC1-XPF complex is responsible for 5'-incision at a dual incision step.
Host Mouse
Species specificity HU
Figures
 |
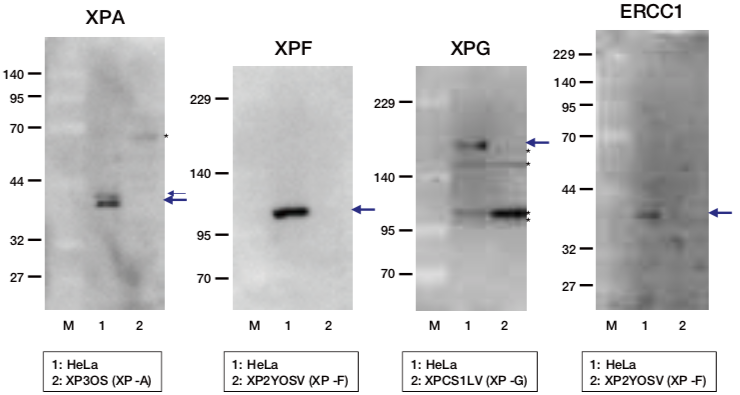 |
| Figure1:Current Model for the Dual Incision Process of NER. | Figure 2: Western blot detection using nucleotide excision repair factor related antibodies. |
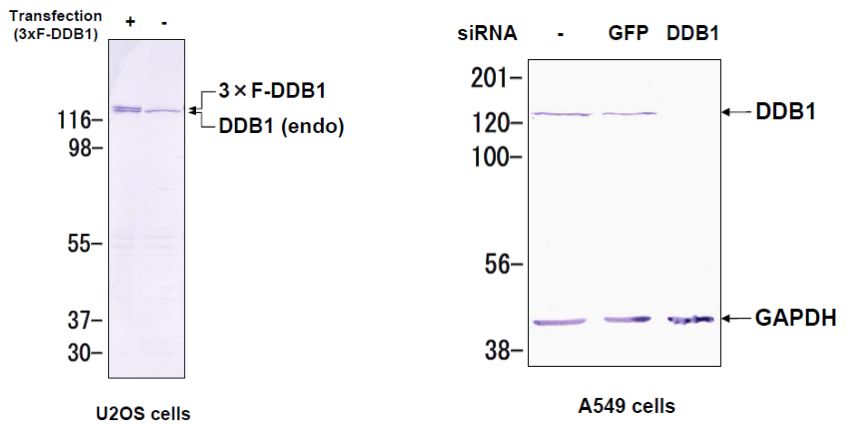
| Product name | Anti DNA Damage-Binding Protein 1 (DDB1) mAb (Clone 43233-3-1) |
| Cat No | CAC-KUP-TM-M05 |
Description
DDB1 was originally identified as a large subunit of damaged DNA-binding protein (DDB), which plays a role in DNA repair. DDB1 also functions as an adaptor molecule of Cul4/DDB1 ubiquitin E3 ligase and participates in various cellular processes.
References:
Wakasugi M, Matsuura K, Nagasawa A, Fu D, Shimizu H, Yamamoto K, Takeda S, Matsunaga T. (2007) DDB1 gene disruption causes a severe growth defect and apoptosis in chicken DT40 cells. Biochem Biophys Res Commun. 364(4):771-7. PMID: 17976535.
Host: MS
Species specificity: HU HAM CHK
 |
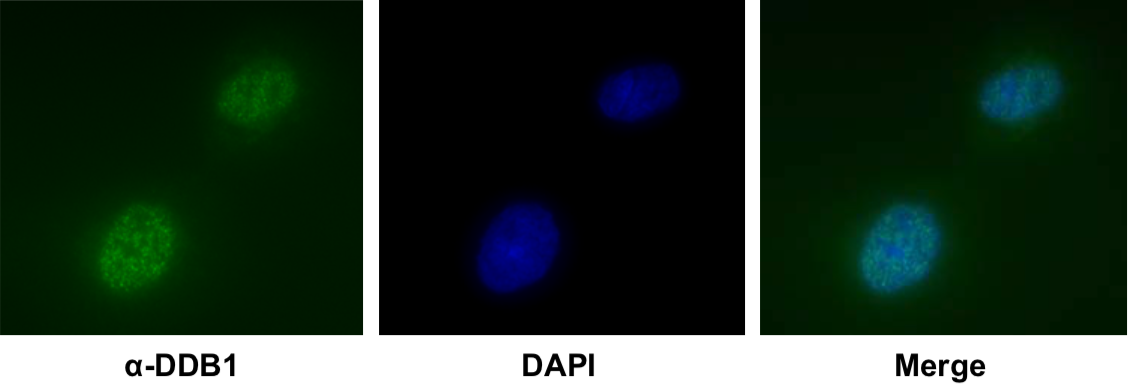 |
| Figure1: Western blot detection using nucleotide excision repair factor related antibodies | Figure 2: Immunofluorescence analysis of U2OS cells with DDB1 mAb clone 43233-3-1 |