Unlock the secrets of protein-protein interactions in cells and tissue with MolBoolean™
MolBoolean™ Mouse/Rabbit
MolBoolean™ is a novel in situ proximity technology developed by Atlas Antibodies that enables the simultaneous detection of both free and interacting fractions for two protein targets.
- Complete spatial quantitative analysis of protein-protein interactions by simultaneous detection of free and interacting proteins.
- Accurate quantification by normalization of interaction data to total target protein levels.
- Biologically relevant data without the need for engineered protein expression.
- 1000-fold increased fluorescence signal by Rolling Circle Amplification, allowing detection and quantification of low abundant proteins.
- Adaptable to different research needs. Universal kit that can be used with the customer’s choice of primary antibodies.
- Validated in both cells and tissue.
 |
|
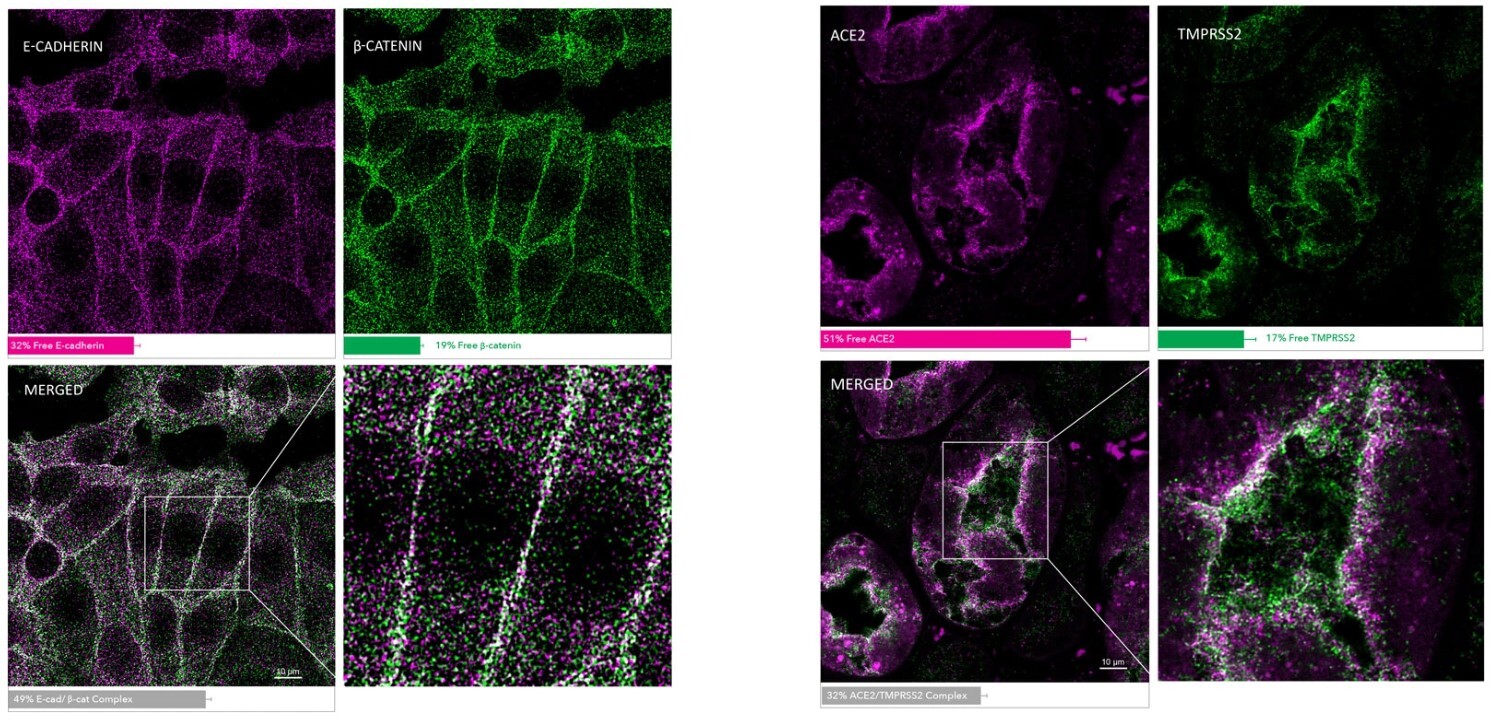
| E-Cadherin/ β-Catenin MolBoolean Staining in MCF7 Cells MolBoolean analysis of the interaction between E-cadherin (cat. AMAb90862, magenta) and β-catenin (cat. HPA029159, green) in MCF7 cells, showing the relative quantification of free versus interacting protein fractions, indicated by the detection of rolling circle products (RCPs) in either one or two fluorescent channels: 32% free E-cadherin (magenta), 19% free β-catenin (green), 49% E-cadherin/β-catenin complex (white). Data is normalized to total target protein levels (total RCPs). |
ACE2/TMPRSS2 MolBoolean Staining in Human Kidney MolBoolean analysis of the interaction between ACE2 (cat. AMAb91259, magenta) and TMPRSS2 (cat. HPA035787, green) in human kidney, showing the relative quantification of free versus interacting protein fractions, indicated by the detection of rolling circle products (RCPs) in either one or two fluorescent channels: 51% free ACE (magenta), 17% free TMPRSS2 (green), 32% ACE/TMPRSS2 complex (white). Data is normalized to total target protein levels (total RCPs). |
Product Description
 MolBoolean™ Mouse/Rabbit is a kit for in situ protein proximity analysis in tissue and cells. The MolBoolean™ assay utilizes a proprietary oligonucleotide setup that enables the simultaneous detection of both free and interacting (~40 nm proximity) fractions for two proteins of interest (protein A, protein B and interaction proteins AB).
MolBoolean™ Mouse/Rabbit is a kit for in situ protein proximity analysis in tissue and cells. The MolBoolean™ assay utilizes a proprietary oligonucleotide setup that enables the simultaneous detection of both free and interacting (~40 nm proximity) fractions for two proteins of interest (protein A, protein B and interaction proteins AB).
MolBoolean™ Mouse/Rabbit should be performed with the user’s primary antibodies of choice (raised in mouse and rabbit). MolBoolean™ relies on anti-mouse and anti-rabbit secondary proximity probes and rolling circle amplification (RCA) as a mean to amplify signal.
A series of molecular steps, performed in the MolBoolean™ assay, incorporates information into the amplified products indicating antibody target engagement of individual versus interacting proteins. This information is converted to fluorescent signals by the binding of detection reporters, allowing visualization on a conventional fluorescent microscope. An image analysis software is provided to segment and differentiate fluorescent signals, enabling the relative quantification of free versus interacting fractions for the two analyzed protein targets in the sample.
| Documents & Links for MolBoolean Mouse/Rabbit Kit (Beyond Proximity Ligation) | |
| Manual | MolBoolean Mouse/Rabbit Kit (Beyond Proximity Ligation) Manual |
| Datasheet | MolBoolean Mouse/Rabbit Kit (Beyond Proximity Ligation) Datasheet |
| MSDS | MolBoolean Mouse/Rabbit Kit (Beyond Proximity Ligation) MSDS |
| Application Note | MolBoolean Mouse/Rabbit Kit (Beyond Proximity Ligation) Application Note |
| Documents & Links for MolBoolean Mouse/Rabbit Kit (Beyond Proximity Ligation) | |
| Manual | MolBoolean Mouse/Rabbit Kit (Beyond Proximity Ligation) Manual |
| Datasheet | MolBoolean Mouse/Rabbit Kit (Beyond Proximity Ligation) Datasheet |
| MSDS | MolBoolean Mouse/Rabbit Kit (Beyond Proximity Ligation) MSDS |
| Application Note | MolBoolean Mouse/Rabbit Kit (Beyond Proximity Ligation) Application Note |
| Citations for MolBoolean Mouse/Rabbit Kit (Beyond Proximity Ligation) – 4 Found |
| Raykova, Doroteya; Kermpatsou, Despoina; Malmqvist, Tony; Harrison, Philip J; Sander, Marie Rubin; Stiller, Christiane; Heldin, Johan; Leino, Mattias; Ricardo, Sara; Klemm, Anna; David, Leonor; Spjuth, Ola; Vemuri, Kalyani; Dimberg, Anna; Sundqvist, Anders; Norlin, Maria; Klaesson, Axel; Kampf, Caroline; Söderberg, Ola. A method for Boolean analysis of protein interactions at a molecular level. Nature Communications. 2022;13(1):4755. PubMed |
| Raykova, Doroteya; Kermpatsou, Despoina; Malmqvist, Tony; Harrison, Philip J; Sander, Marie Rubin; Stiller, Christiane; Heldin, Johan; Leino, Mattias; Ricardo, Sara; Klemm, Anna; David, Leonor; Spjuth, Ola; Vemuri, Kalyani; Dimberg, Anna; Sundqvist, Anders; Norlin, Maria; Klaesson, Axel; Kampf, Caroline; Söderberg, Ola. Author Correction: A method for Boolean analysis of protein interactions at a molecular level. Nature Communications. 2023;14(1):5450. PubMed |
| Rivas-Santisteban, Rafael; Rico, Alberto José; Muñoz, Ana; Rodríguez-Pérez, Ana I; Reyes-Resina, Irene; Navarro, Gemma; Labandeira-García, José Luis; Lanciego, José Luis; Franco, Rafael. Boolean analysis shows a high proportion of dopamine D(2) receptors interacting with adenosine A(2A) receptors in striatal medium spiny neurons of mouse and non-human primate models of Parkinson's disease. Neurobiology Of Disease. 2023;188:106341. PubMed |
| Kotliar, Ilana B; Bendes, Annika; Dahl, Leo; Chen, Yuanhuang; Saarinen, Marcus; Ceraudo, Emilie; Dodig-Crnković, Tea; Uhle'n, Mathias; Svenningsson, Per; Schwenk, Jochen M; Sakmar, Thomas P. Expanding the GPCR-RAMP interactome. Biorxiv : The Preprint Server For Biology. 2023; 38045268 PubMed |





