Exosomes are small vesicles with diameters of 30 to 200 nm that are secreted from almost all cells that make up the body, and are present in all body fluids such as blood and urine (1)-(5) . They are
also secreted into the culture supernatant of animal cells in vitro . Like cells, exosomes are surrounded by a lipid bilayer membrane, with membrane proteins present on their surface and proteins and microRNAs contained inside. It is thought that exosomes are responsible for intercellular communication through the function of these proteins and microRNAs in target cells that have taken up exosomes (6) . One of the structural characteristics of exosomes is the tetraspanin family present on their surface. CD9, CD63, and CD81 are member molecules of this family and are also surface markers for exosomes (7) .
Methods for quantifying exosomes include the amount of protein contained in exosomes and particle analysis using the nanotracking method (8) , but these methods require exosomes to be purified first by ultracentrifugation or other methods. There are very limited means to directly quantify exosomes in body fluids or cell culture fluids, and no general method has been developed to date.
This kit uses sandwich ELISA with high-performance antibodies against the exosome markers CD9, CD63, or CD81, respectively, to relatively quantify exosomes that have CD9, CD63, or CD81 molecules on their surface.
Features
- Direct quantification of exosomes contained in blood samples, cell culture supernatants, etc.
- No special equipment is required; measurements can be performed with a standard plate reader.
- Instead of using exosomes as a standard reagent, which lack storage stability, we use 200 nm beads immobilized with CD9, CD63, or CD81 to ensure stability and reproducibility.
- Relative quantification of each sample is possible by normalizing with standard beads CD9, CD63, or CD81 that mimic exosome structure
- Exosomes are captured with immobilized CD9, CD63 or CD81 antibodies and detected with HRP-conjugated CD9, CD63 or CD81 antibodies.
Kit components
- Anti-CD9 *1 antibody immobilized 96 well plate 8-well x 12 strips 1 sheet
- CD9 *1 Standard beads 200μL x 1
- Assay buffer 25mL x 1
- Washing buffer (10X) 25mL x 1
- HRP labeled anti-CD9 *1 antibody (500X) 20μL 1 bottle
- Substrate solution 12mL x 1
- Stop solution (2N H 2 SO 4 ) 6mL 1 bottle
- Plate seal x 2
*1: Part number HAK-HEL6363-1 is CD63 Part number HAK-HEL8181-1 is CD81
*2: n=2, calibration curve 4 times
Example data
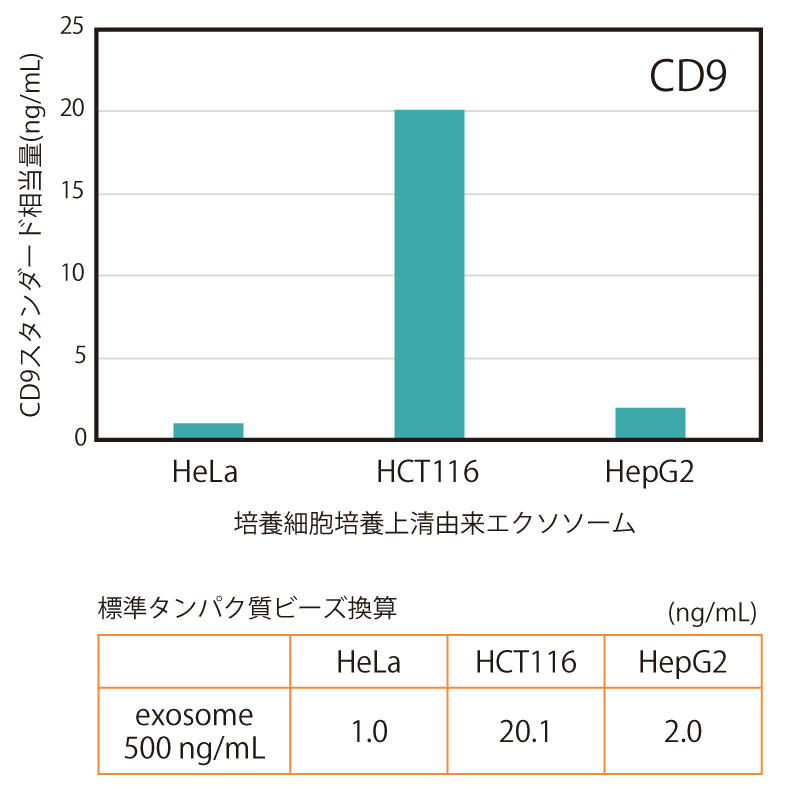
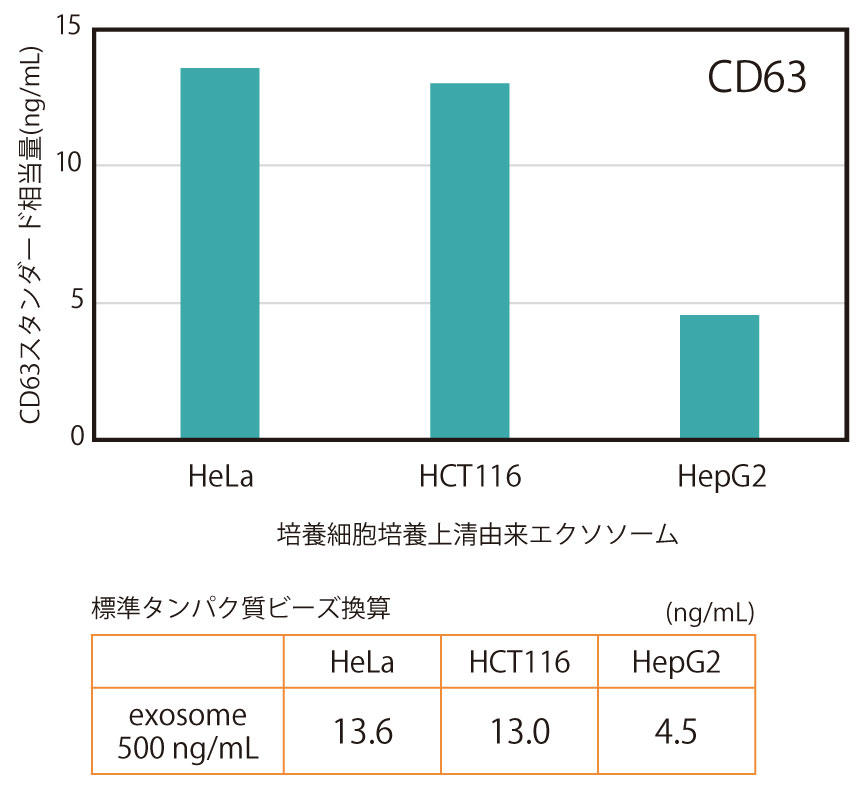
Exosomes purified by ultracentrifugation from the culture supernatant (serum-free) of three types of human cancer cell lines (HeLa, HCT116, HepG2) were diluted to a concentration of 500 ng/mL and measured by CD9/CD9 ELISA (Figure 1) or CD63/CD63 ELISA (Figure 2).
Quantitative values were plotted against a standard curve using standard beads immobilized with CD9 (or CD63) protein, and expressed as the equivalent amount of standard beads (ng/mL).
As a result, it was found that the relative amounts of CD9 and CD63 on the surface of the cells varied depending on the cell type.
Standardization of measurements
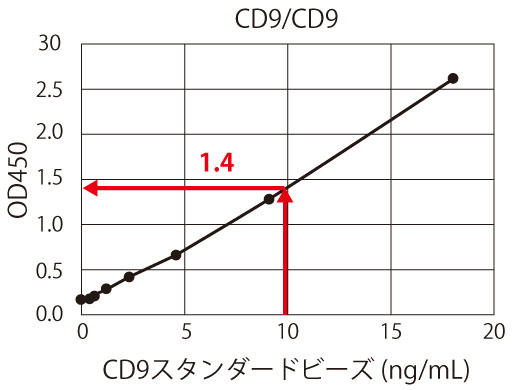
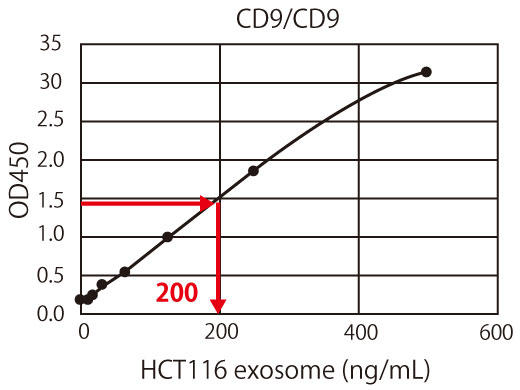
Figure 3. Overview of normalization and relative quantification using CD9 standard beads
The graph obtained from the measurement results of the CD9 standard is used as the standard curve. For example, if 10 ng/mL is 1 U/mL, the OD 450 measurement value is approximately 1.4 (Figure 3(a)). The OD 450 measurement value of 1.4 for the HCT116-derived exosome sample corresponds to the protein equivalent of approximately 200 ng/mL of exosomes, as shown in Figure 3(b). In other words, approximately 200 ng/mL of HCT116 cell-derived exosomes can be considered as 1/mL of CD9-positive exosomes. In this way, the exosome measurements between different samples or different experiments can be standardized and corrected by showing all the measurements in units, making it possible to directly compare the amount of exosomes in the sample.
- J. Skog, T. Wurdinger, S. van Rijn, DH Meijer, L. Gainche, M. Sena-Esteves, WT Jr. Curry, BS Carter, AM Krichevsky and XO Breakefield: Nat Cell Biol ., 10 , 1470 (2008 ).
- T. Pisitkun, RF Shen and MA Knepper: Proc Natl Acad Sci USA ., 101 , 13368 (2004).
- S. Runz, S. Keller, C. Rupp, A. Stoeck, Y. Issa, D. Koensgen, A. Mustea, J. Sehouli, G. Kristiansen and P. Altevogt: Gynecol Oncol ., 107 , 563 (2007) .
- S. Keller, AK Konig, F. Marme, S. Runz, S. Wolterink, D. Koensgen, A. Mustea, J. Sehouli and P. Altevogt: Cancer Lett ., 278 , 73 (2009).
- C. Lasser, V. Seyed AlikhS. Gabrielsson, J. Lotvall and H. Valadi: J Transl Med ., 9 , 9 (2011).
- Y. Naito, Y. Yoshioka, Y. Yamamoto and T. Ochiya: Cell Mol Life Sci ., 74 , 697 (2017).
- A. Zoraida and M. Yanez-Mo: Front Immunol ., 5 , 442 (2014).
- V. Filipe, A. Hawe and W. Jiskoot: Pharm Res ., 27 , 796 (2010).
| Product Specifications | |
| Reactivity | Human |
| Documents & Links for CD9/CD9 Exosome ELISA Kit (Human) | |
| Datasheet | CD9/CD9 Exosome ELISA Kit Datasheet |
| Documents & Links for CD9/CD9 Exosome ELISA Kit (Human) | |
| Datasheet | CD9/CD9 Exosome ELISA Kit Datasheet |