 CAC Antibody Collection
CAC Antibody Collection
The antibodies on this page are part of Cosmo Bio's exclusive CAC Collection. For many many thousands of other antibodies from many different makers, use our Search the Store function and our Explore Products drop down menu.
Epigenetics and chromatin
Histone H3 variants / Post-translationally-modified histone H3 / Chromatin structure modifiers / Drosophila chromatin
| Epigenetics and chromatin: Histone H3 variants | |||
| Product name (click for order info) | Cat No (click for datasheet) |
Host | Species specificity |
| Anti Histone H3.1(HIST1H3A) / H3.2 (HIST1H3B) mAb (Clone 6G3C7) | CAC-CEC-005 | RT | HU MS RT MKY HAM |
| Anti Histone H3.1(HIST1H3A) / H3.2 (HIST1H3B) mAb (Clone 1D4F2) | CAC-CEC-006 | MS | HU MS MKY RT HAM CAN |
| Anti Histone H3.3 (H3F3A) mAb (Clone 6C4A3) | CAC-CEC-007 | RT | HU MS RT MKY CAN |
| Anti Histone H3.3 (H3F3A) mAb (Clone 4H2D7) | CAC-CEC-008 | RT | HU MS MKY RT HAM |
| Epigenetics and chromatin: Post-translationally-modified histone H3 | |||
| Product name (click for order info) | Cat No (click for datasheet) |
Host | Species specificity |
| Anti Histone H3.1 (HIST1H3A) S10ph mAb (Clone 6G8B7) | CAC-CE-001 | RT | HU MS RT MKY HAM |
| Anti Histone H3.1 (HIST1H3A) T11ph mAb (Clone 6G12C5) | CAC-CE-002 | RT | HU MS RT MKY HAM |
| Anti Histone H3.1 (HIST1H3A) T32ph mAb (Clone 6C7G12) | CAC-CE-003 | RT | HU MS RT MKY HAM |
| Anti Histone H3.1 (HIST1H3A) K9Ac mAb (Clone 2G1F9) | CAC-CE-004 | RT | HU MS RT MKY HAM |
| Epigenetics and chromatin: Drosophila chromatin | |||
| Product name (click for order info) | Cat No (click for datasheet) |
Host | Species specificity |
| Anti Drosophila FACT Complex Subunit Ssrp1 (dSSRP1) pAb (Rabbit, Antiserum) | CAC-NIG-L1-SHA1 | RAB | Drosophila |
| Anti Drosophila FACT Complex Subunit Spt16 (dSPT16/DRE4) pAb (Rabbit, Antiserum) | CAC-NIG-L1-SHA2 | RAB | Drosophila |
Histone H3 variants
H3.3 differs from the canonical replication-coupled H3 histones (H3.1/H3.2 in mammals) by only four to five amino acids, respectively, but has distinct expression patterns and chromatin incorporation mechanisms. Canonical H3 variants are expressed only during S phase, and their incorporation is mediated by the CAF-1 chromatin assembly complex (Smith and Stillman 1989; Verreault et al.1996; Tagami et al. 2004). In contrast, H3.3 is expressed throughout the cell cycle and is incorporated via a number of different pathways, including the CHD1 and ATRX chromatin remodeling complexes and the HIRA chaperone complex (Tagami et al. 2004; Konev et al. 2007; Draneet al. 2010; Lewis et al. 2010). H3.3 is unevenly distributed in the genome, suggesting that it may have diverse roles. It is enriched in transcribed genes, enhancers, regulatory elements, and also heterochromatic repeats, including telomeres and pericentromeric regions (Wirbelauer et al. 2005; Loyola and Almouzni 2007; Goldberg et al. 2010; Szenker et al.2011). A study with mouse embryonic stem (ES) cells showed that the HIRA complex deposits H3.3 in transcribed genes, while the ATRX complex delivers H3.3 to telomeres and pericentromeric repeats (Goldberg et al.2010). H3.3 has been implicated in the epigenetic regulation of diverse biological processes. When incorporated into a nucleosome, H3.3 accumulates modifications associated with gene activation or open chromatin—including methylation on K4, K36, and K79 and acetylation on K9 and K14—and is deprived of suppressive modifications, including K9 and K27 methylation (McKittrick et al. 2004; Loyola and Almouzni 2007). H3.3-containing nucleosomes are intrinsically unstable and promote gene activation, especially with histone H2A.Z co-incorporation (Jin and Felsenfeld 2007; Jin et al. 2009; Chen et al. 2013). H3.3 maintains the somatic epigenetic memory in nuclear transferred Xenopus nuclei (Ng and Gurdon 2008) and supports differentiation of cultured murine muscle precursor cells (Harada et al. 2012). HIRA-dependent incorporation of H3.3 is also required for the reprogramming of nuclear transferred murine nuclei (Jullien et al. 2012; Wen et al. 2014), DNA replication and rRNA transcription in mouse zygotes (Lin et al. 2014), transcriptional recovery after genomic DNA damage in human cancer cell lines (Adam et al. 2013), and Polycomb-repressive complex 2 recruitment to developmentally regulated bivalent loci in mouse ES cells (Banaszynski et al. 2013). Furthermore, somatic mutations in H3.3-encoding genes cause various types of cancer in humans, underscoring its role in maintaining tissue homeostasis (Schwartzentruber et al. 2012; Behjati et al. 2013). (From: Jang et al. Histone H3.3 maintains genome integrity during mammalian development. Genes & Development 2019; 29:1377–1392)| Product name | Anti Histone H3.1(HIST1H3A) / H3.2 (HIST1H3B) mAb (Clone 6G3C7) | |
| Cat No | CAC-CEC-005 | |
| Description | Nucleosomes are composed of four different histone proteins, designated H2A, H2B, H3, and H4. In humans, five variants of histone H3 are reported: H3.1, H3.2, H3.3, H3t, and CENP-A. The two major Histone H3 variants (H3.1 and H3.3) display distinct genomic localization patterns in eukaryotes. Deposition of Histone H3.1 is associated with DNA synthesis during DNA replication and possibly DNA repair, while Histone H3.3 is incorporated independently of DNA synthesis and is the predominant form of H3 found in non-dividing cells. Recently, it was shown that a genomic gene cluster regulating skeletal myogenesis is marked by H3.3 protein prior to cellular muscle formation and that H3.3 marking of this region enables myogenic gene activation. These results suggest that monitoring H3.3 marking at specific loci may be useful in the prediction of cell fate. Hence, this new histone H3 variant monoclonal antibody has great utility for dissecting the functional significance of H3 variants and the molecular mechanisms associated with their deposition. References: 1) Hake SB et al., Proc Natl Acad Sci USA (2006) 103 (17):6428-35. PMID: 16571659 2) Harada et al., EMBOJ (2012) 31(13):2994-3007. PMID:225 |
|
| Host | Rat | |
| Species specificity | HU MS RT MKY HAM | |
| Figure 1 |  |
|
| The composition of Histone H3 variant peptides and reactivity using histone H3.1/H3.2 antibody 6G3C7. | ||
| Figure 2 |  |
|
| Western blot analysis of recombinant proteins and mammalian cell extracts using Histone H3.1/H3.2 antibody 6G3C7. | ||
| Product name | Anti Histone H3.1(HIST1H3A) / H3.2 (HIST1H3B) mAb (Clone 1D4F2) | |
| Cat No | CAC-CEC-006 | |
| Description | Nucleosomes are composed of four different histone proteins, designated H2A, H2B, H3, and H4. In humans, five variants of histone H3 are reported: H3.1, H3.2, H3.3, H3t, and CENP-A. The two major Histone H3 variants (H3.1 and H3.3) display distinct genomic localization patterns in eukaryotes. Deposition of Histone H3.1 is associated with DNA synthesis during DNA replication and possibly DNA repair, while Histone H3.3 is incorporated independently of DNA synthesis and is the predominant form of H3 found in non-dividing cells. Recently, it was shown that a genomic gene cluster regulating skeletal myogenesis is marked by H3.3 protein prior to cellular muscle formation and that H3.3 marking of this region enables myogenic gene activation. These results suggest that monitoring H3.3 marking at specific loci may be useful in the prediction of cell fate. Hence, this new histone H3 variant monoclonal antibody has great utility for dissecting the functional significance of H3 variants and the molecular mechanisms associated with their deposition. References: 1) Hake SB et al., Proc Natl Acad Sci USA (2006) 103 (17):6428-35. PMID: 16571659 2) Harada et al., EMBOJ (2012) 31(13):2994-3007. PMID:225 |
|
| Host | Mouse | |
| Species specificity | HU MS RT MKY HAM | |
| Figure 1 | 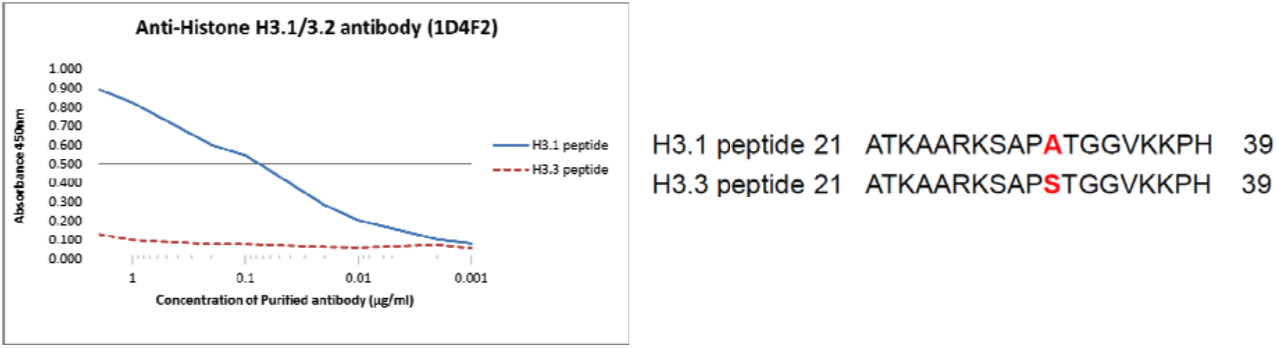 |
|
| The composition of Histone H3 variants peptides and the reactivity of Histone H3.1/H3.2 antibody 1D4F2. | ||
| Figure 2 | 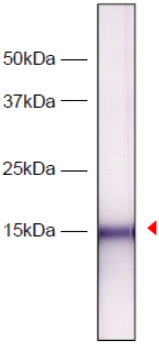 |
|
| Western blot analysis of HeLa cell extracts using Histone H3.1/H3.2 antibody 1D4F2. | ||
| Figure 3 | 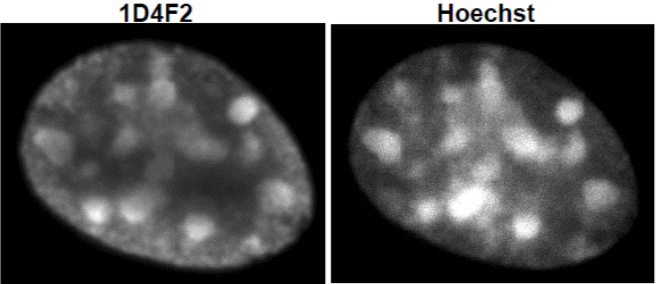 |
|
| Immunocytochemical analysis of NIH3T3 cells using Histone H3.1/H3.2 antibody 1D4F2. | ||
| Product name | Anti Histone H3.3 (H3F3A) mAb (Clone 6C4A3) | |
| Cat No | CAC-CEC-007 | |
| Description | Nucleosomes are composed of four different histone proteins, designated H2A, H2B, H3, and H4. In humans, five variants of histone H3 are reported: H3.1, H3.2, H3.3, H3t, and CENP-A. The two major Histone H3 variants (H3.1 and H3.3) display distinct genomic localization patterns in eukaryotes. Deposition of Histone H3.1 is associated with DNA synthesis during DNA replication and possibly DNA repair, while Histone H3.3 is incorporated independently of DNA synthesis and is the predominant form of H3 found in non-dividing cells. Recently, it was shown that a genomic gene cluster regulating skeletal myogenesis is marked by H3.3 protein prior to cellular muscle formation and that H3.3 marking of this region enables myogenic gene activation. These results suggest that monitoring H3.3 marking at specific loci may be useful in the prediction of cell fate. Hence, this new histone H3.3 variant monoclonal antibody is expected to be useful in the field of regenerative medicine. References: 1) Hake SB et al., Proc Natl Acad Sci USA (2006) 103 (17):6428-35. PMID: 16571659 2) Harada et al., EMBOJ (2012) 31(13):2994-3007. PMID:225 |
|
| Host | Rat | |
| Species specificity | HU MS RT MKY CAN | |
| Figure 1 | 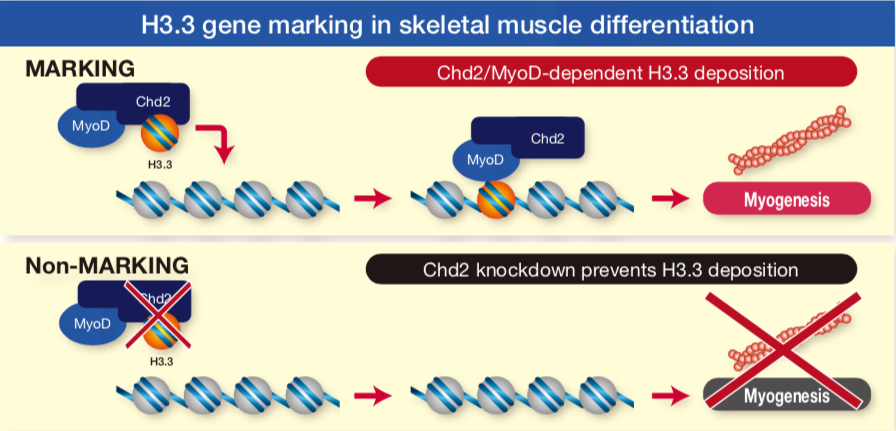 |
|
| These H3 variant antibodies were essential tools in a first of kind study showing that differentiation specific genes are marked for lineage specific expression by the deposition of Histone H3.3 at the onset of differentiation signaling (Ref. 2). | ||
| Figure 2 | 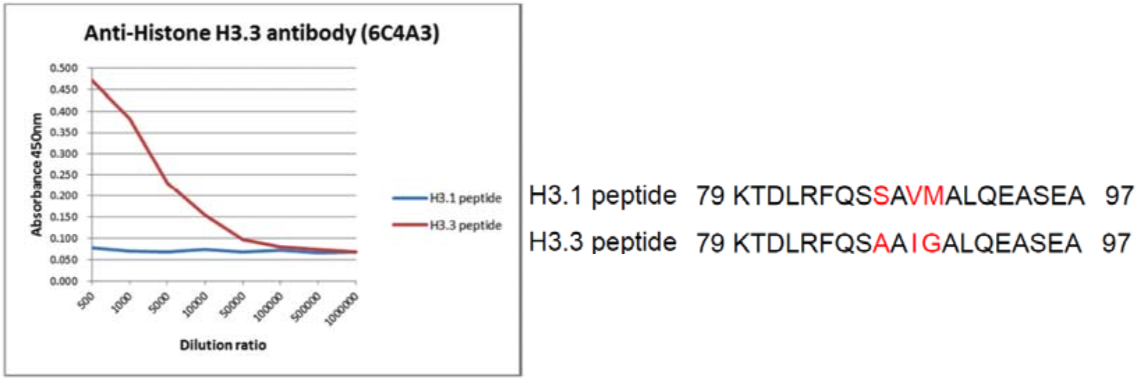 |
|
| The composition of histone H3 variant peptides and reactivity of Histone H3.3 antibody 6C4A3. | ||
| Figure 3 | 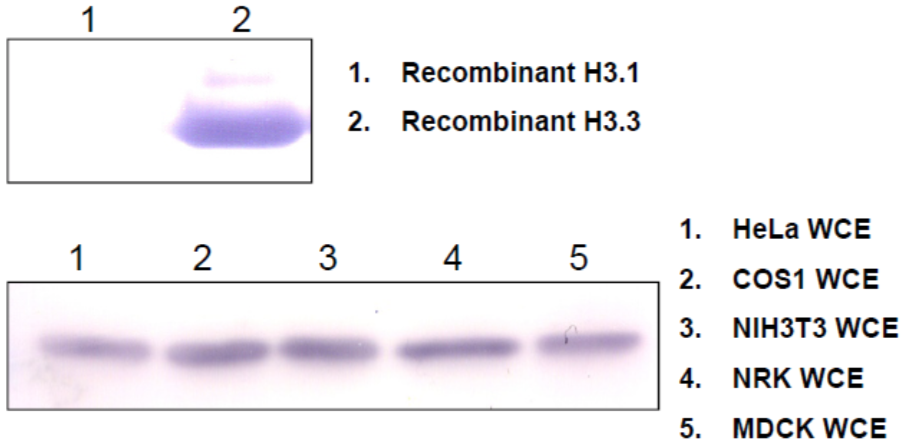 |
|
| Western blot analysis of recombinant proteins and mammalian cell extracts using Histone H3.3 antibody 6C4A3. | ||
| Product name | Anti Histone H3.3 (H3F3A) mAb (Clone 4H2D7) | |
| Cat No | CAC-CEC-008 | |
| Description | Nucleosomes are composed of four different histone proteins, designated H2A, H2B, H3, and H4. In humans, five variants of histone H3 are reported: H3.1, H3.2, H3.3, H3t, and CENP-A. The two major Histone H3 variants (H3.1 and H3.3) display distinct genomic localization patterns in eukaryotes. Deposition of Histone H3.1 is associated with DNA synthesis during DNA replication and possibly DNA repair, while Histone H3.3 is incorporated independently of DNA synthesis and is the predominant form of H3 found in non-dividing cells. Recently, it was shown that a genomic gene cluster regulating skeletal myogenesis is marked by H3.3 protein prior to cellular muscle formation and that H3.3 marking of this region enables myogenic gene activation. These results suggest that monitoring H3.3 marking at specific loci may be useful in the prediction of cell fate. Hence, this new histone H3.3 variant monoclonal antibody is expected to be useful in the field of regenerative medicine. References: 1) Hake SB et al., Proc Natl Acad Sci USA (2006) 103 (17):6428-35. PMID: 16571659 2) Harada et al., EMBOJ (2012) 31(13):2994-3007. PMID:225 |
|
| Host | Rat | |
| Species specificity | HU MS MKY RT HAM | |
| Figure 1 | 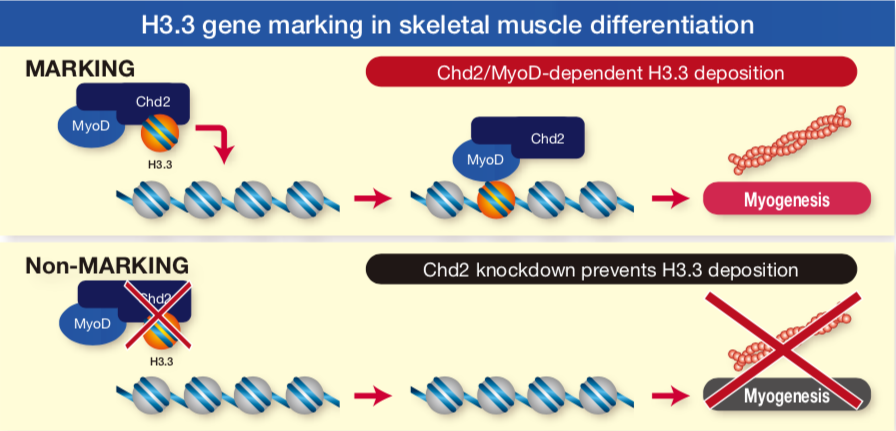 |
|
| These H3 variant antibodies were essential tools in a first of kind study showing that differentiation specific genes are marked for lineage specific expression by the deposition of Histone H3.3 at the onset of differentiation signaling (Ref. 2). | ||
| Figure 2 | 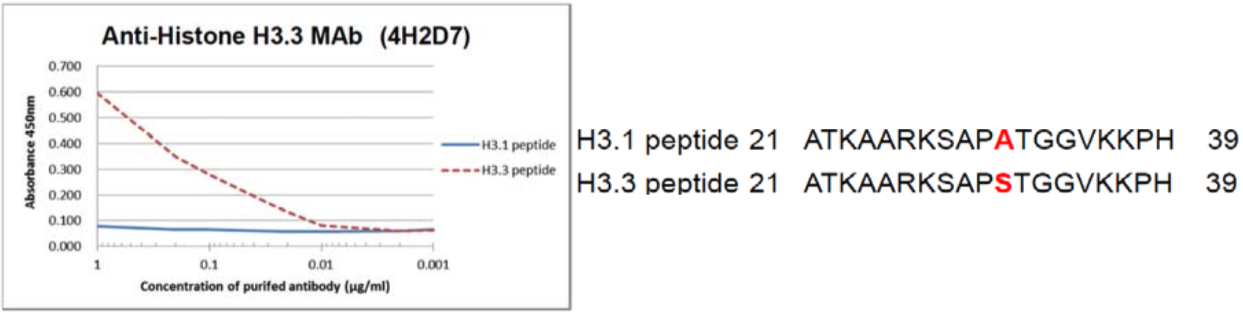 |
|
| The composition of Histone H3 variant peptides and reactivity of Histone H3.3 antibody 4H2D7. | ||
| Figure 3 | 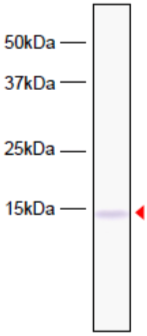 |
|
| Western blot analysis of HeLa cell extracts using Histone H3.3 antibody 4H2D7. | ||
| Figure 4 | 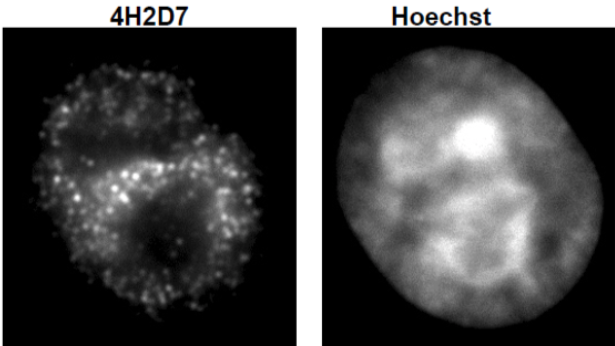 |
|
| Immunocytochemical analysis of HeLa cells using Histone H3.3 antibody 4H2D7. | ||
| Post-translationally-modified histone H3 |
||
| Similarly to DNA methylation, posttranslational histone modifications do not affect DNA nucleotide sequence but can modify its availability to the transcriptional machinery. Several types of histone modifications are known, amongst which acetylation, methylation, phosphorylation, and ubiquitination are the best studied and most important in terms of the regulation of chromatin structure and (transcriptional) activity. In general, histone modifications are catalyzed by specific enzymes that act, predominantly, but not exclusively (e.g. some types of histone phosphorylation), at the histone N-terminal tails involving amino acids such as lysine or arginine as well as serine, threonine, tyrosine, etc. Histone acetylation usually leads to higher gene expression. This may not always be the case for histone H4. Histone methylation in turn has either transcriptionally permissive or repressive character, depending on the location of targeted amino acid residues in the histone tail and/or the number of modifying (e.g. methyl) groups added. [adapted from: Alaskhar Alhamwe et al. Allergy Asthma Clin Immunol (2018) 14:39 https://doi.org/10.1186/s13223-018-0259-4] | ||
| Product name | Anti Histone H3.1 (HIST1H3A) S10ph mAb (Clone 6G8B7) | |
| Cat No | CAC-CE-001 | |
| Description | Post-translation modifications of histones modulate the accessibility and transcriptional competence of specific chromatin regions within the eukaryotic genome. Phosphorylation of histone H3 is unique in the sense that it associates on one hand with open chromatin during gene activation and marks on the other hand highly condensed chromatin during mitosis. References: Strahl and Allis, (2000) Nature. 403(6765):41-5. PMID: 10638745. |
|
| Host | RT | |
| Species specificity | HU MS RT MKY HAM | |
| Figure 1 | 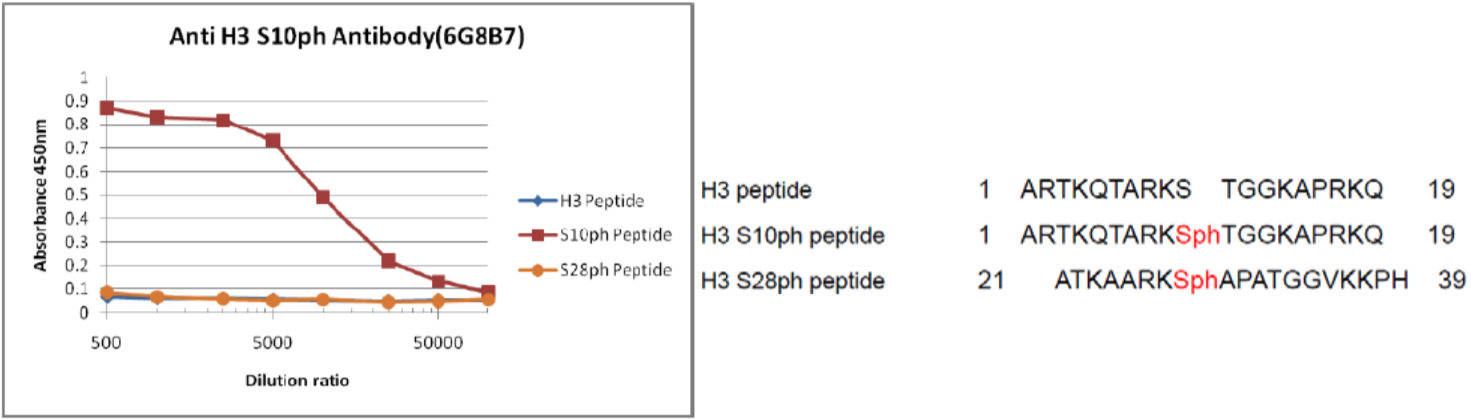 |
|
| Reactivity of histone H3 S10ph antibody 6G8B7 against histone H3 peptides. | ||
| Figure 2 | 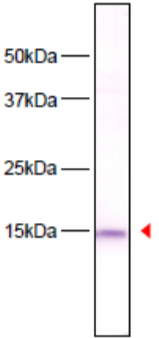 |
|
| Immunoblot analysis of HeLa cell extracts using histone H3 S10ph antibody 6G8B7. | ||
| Figure 3 | 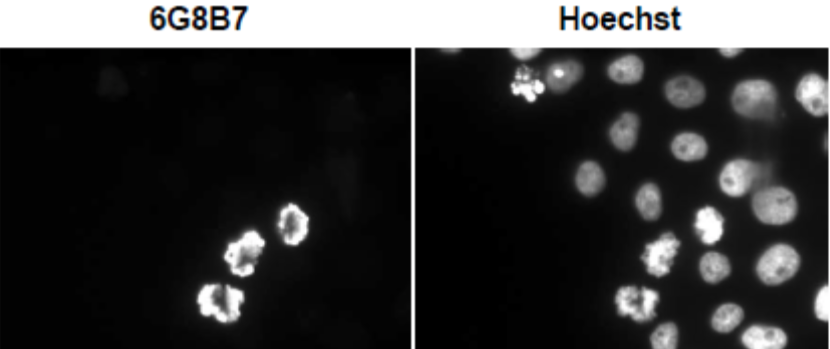 |
|
| Immunocytochemical analysis of HeLa Cell using histone H3 S10ph antibody 6G8B7. | ||
| Product name | Anti Histone H3.1 (HIST1H3A) T11ph mAb (Clone 6G12C5) | |
| Cat No | CAC-CE-002 | |
| Description | Post-translation modifications of histones modulate the accessibility and transcriptional competence of specific chromatin regions within the eukaryotic genome. Phosphorylation of histone H3 Threonine 11 (H3-T11ph) occurs throughout the cell cycle and is Chk1 dependent. It has been reported that DNA damage rapidly reduces H3-T11 phosphorylation. References: Shimada M, et al. (2008) Cell. 132(2):221-32. PMID: 18243098. |
|
| Host | RT | |
| Species specificity | HU MS RT MKY HAM | |
| Figure 1 | 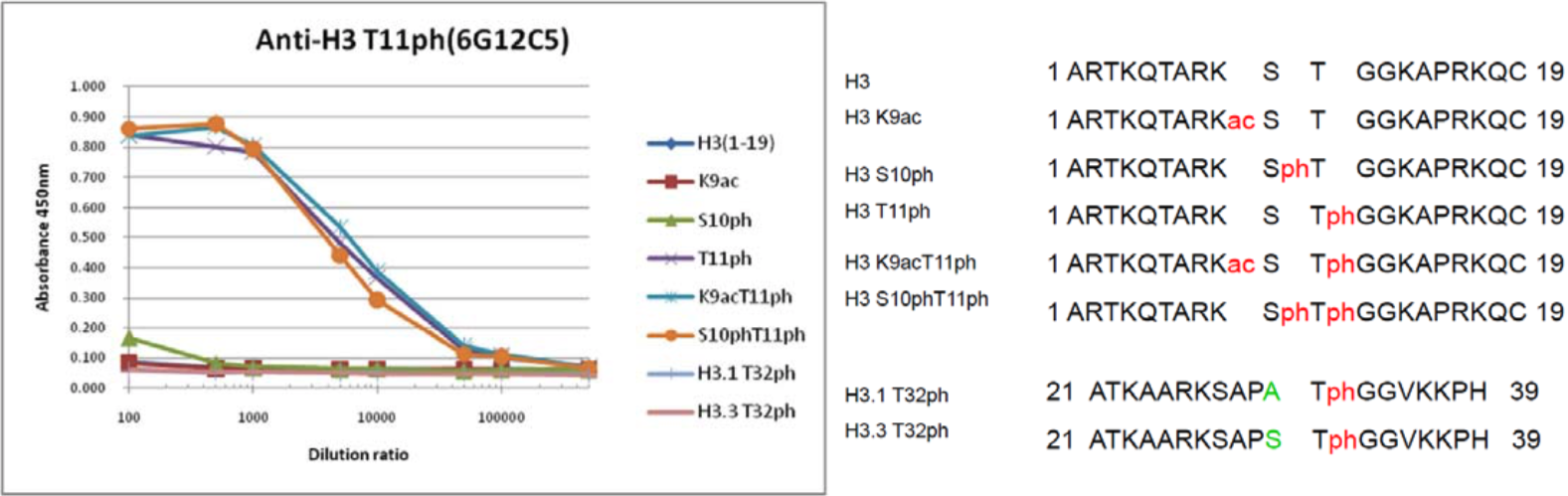 |
|
| The composition of histone H3 peptides and the reactivity of histone H3 T11ph antibody 6G12C5. | ||
| Figure 2 | 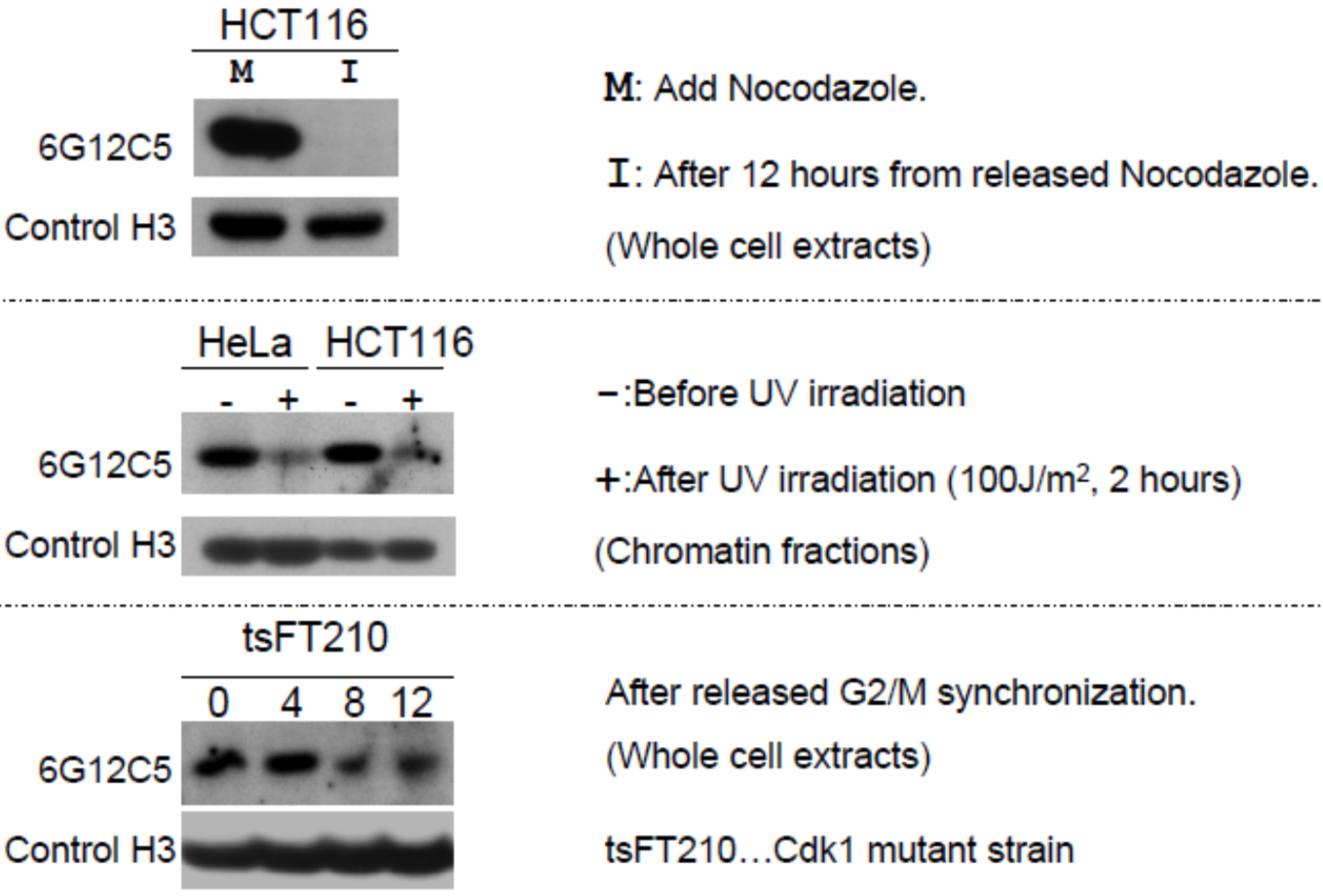 |
|
| Immunoblot analysis of treated-cell extracts using histone H3 T11ph antibody 6G12C5. | ||
| Figure 3 | 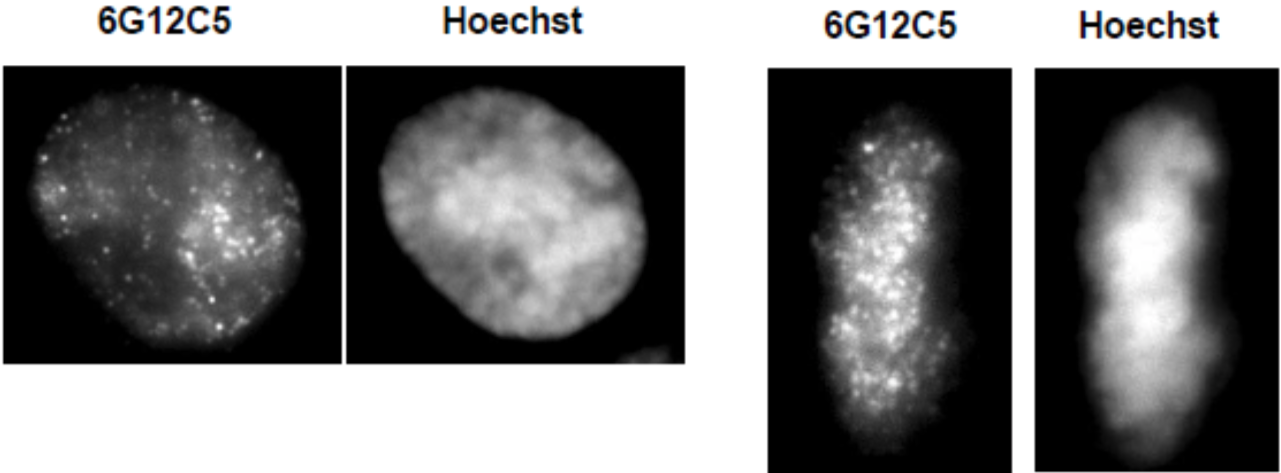 |
|
| Immunocytochemical analysis of HeLa cells using histone H3 T11ph antibody 6G12C5 (left: interphase, right: metaphase). | ||
| Product name | Anti Histone H3.1 (HIST1H3A) T32ph mAb (Clone 6C7G12) | |
| Cat No | CAC-CE-003 | |
| Description | Post-translation modifications of histones modulate the accessibility and transcriptional competence of specific chromatin regions within the eukaryotic genome. Phosphorylation of histone H3 is unique in the sense that it associates on one hand with open chromatin during gene activation and marks on the other hand highly condensed chromatin during mitosis. References: Strahl and Allis. (2000) Nature. 403(6765):41-5. PMID: 10638745. |
|
| Host | RT | |
| Species specificity | HU MS RT MKY HAM | |
| Figure 1 |  |
|
| The composition of Histone H3 peptides and the reactivity of histone H3 T32ph antibody 6C7G12. | ||
| Figure 2 | 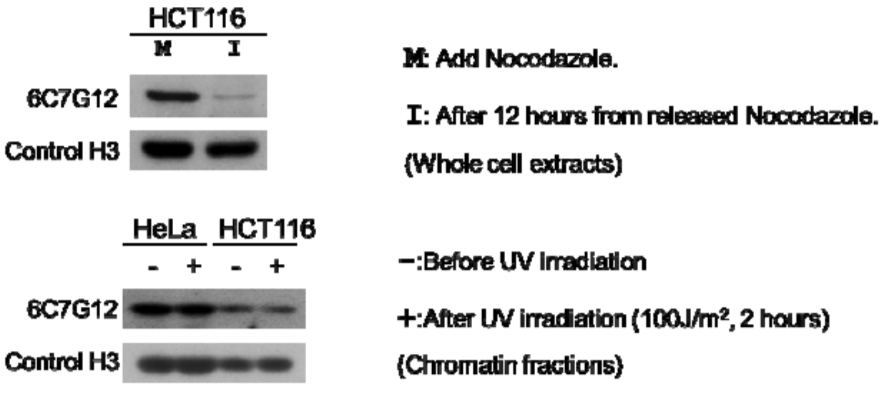 |
|
| Immunoblot analysis of treated-cell extracts using histone H3 T32ph antibody 6C7G12. | ||
| Figure 3 |  |
|
| Immunocytochemical analysis of HeLa cells using histone H3 T32ph antibody 6C7G12. | ||
| Product name | Anti Histone H3.1 (HIST1H3A) K9Ac mAb (Clone 2G1F9) | |
| Cat No | CAC-CE-004 | |
| Description | Post-translational modifications of histones modulate the accessibility and transcriptional competence of specific chromatin regions within the eukaryotic genome. Histone H3 is primarily acetylated at lysines 9, 14, 18, and 23. Acetylation at lysine 9 appears to have a dominant role in histone deposition and chromatin assembly. References: Strahl and Allis. (2000) Nature. 403(6765):41-5. PMID: 10638745. |
|
| Host | RT | |
| Species specificity | HU MS RT MKY HAM | |
| Figure 1 | 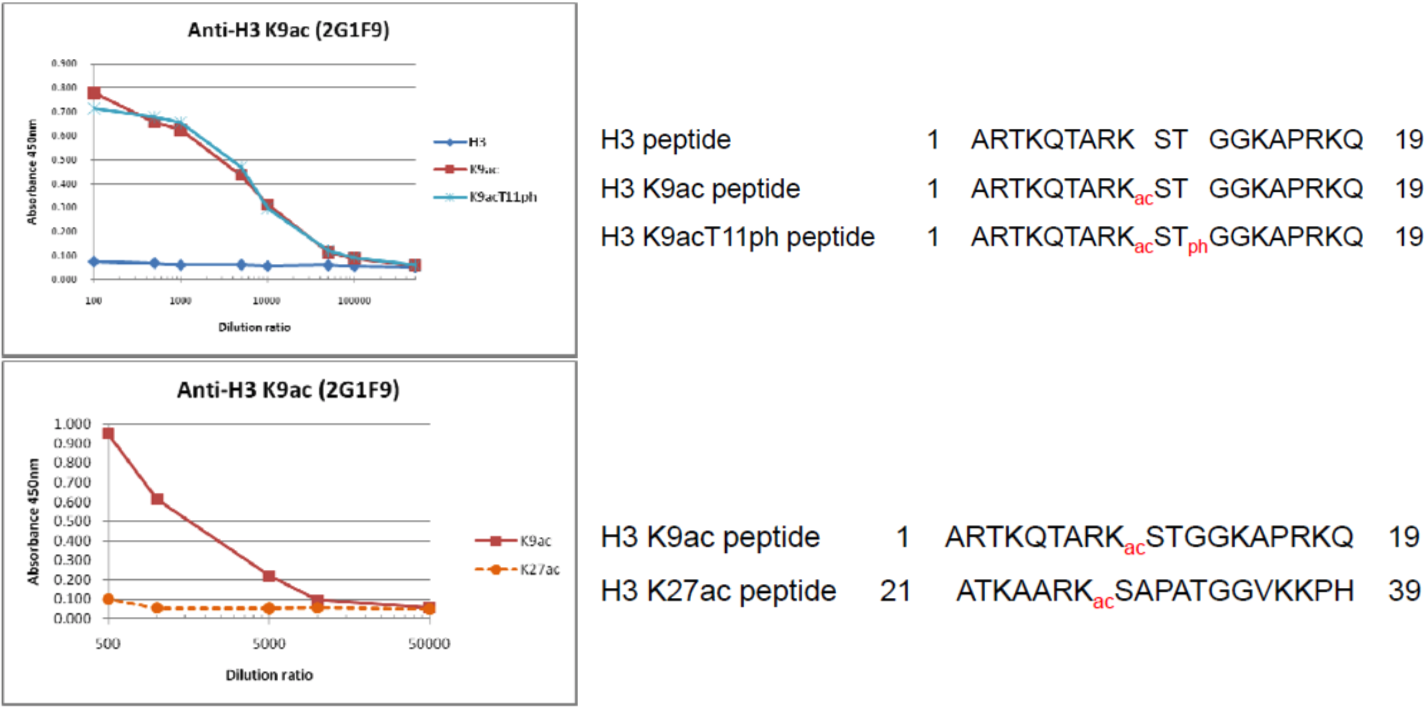 |
|
| The composition of histone H3 peptides and the reactivity of histone H3 K9Ac antibody 2G1F9. | ||
| Figure 2 | 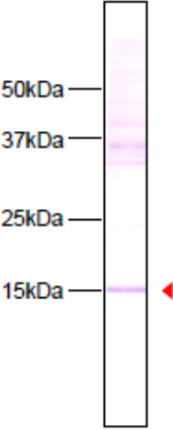 |
|
| Immunoblot analysis of HeLa cell extracts using histone H3 K9ac antibody 2G1F9. | ||
| Figure 3 | 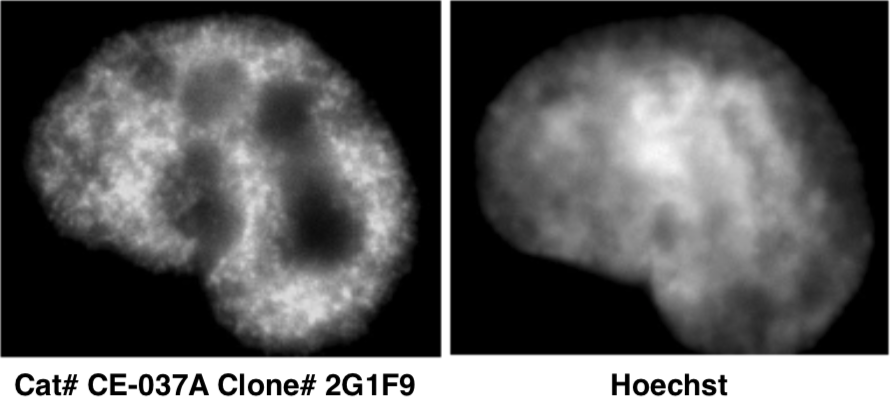 |
|
| Immunocytochemical analysis of HeLa cells using histone H3 K9ac antibody 2G1F9. | ||
| Chromatin structure modifiers |
||
| The accessibility of genomic binding sites for transcription factors and Pol II at enhancer and promoter regions is essential for efficient gene expression. Chromatin accessibility is controlled by DNA methylation, histone modifications and chromatin modifiers, but also chromatin remodeling and 3D chromatin organization are relevant in this process. Chromatin remodelers are multi-protein complexes that use the energy of ATP hydrolysis, in order to slide, exchange or remove nucleosomes. Chromatin remodelers make promoter and enhancer regions either more or less accessible to the transcriptional apparatus, thereby allowing transcription factors to activate or repress, respectively, the transcription of their target genes. Thus, nucleosome occupancy and composition are tailored genome-wide by specialized remodelers. CTCF and cohesin are the main proteins organizing chromatin into architectural loops. Within these TADs, regulatory loops between enhancer and TSS regions activate their target genes, i.e. the reach of an enhancer is primarily restricted to the TAD in which it resides. Disease-associated mutations in chromatin loop anchors and rearrangements of TADs allow a mechanistic understanding of dys-regulated target genes. Some of the thousands of long ncRNAs encoded by the human genome are implicated in chromatin regulation and organization. Mechanistic insight into the X chromosome inactivating ncRNA Xist outlines the principles how these molecules regulate gene expression by coordinating regulatory proteins, localizing to target loci and shaping 3D chromatin organization. [From: Carlberg C., Molnár F. (2018) Chromatin Remodelers and Organizers. In: Human Epigenomics. Springer, Singapore] | ||
| Product name | Anti Developmental Pluripotency-Associated Protein 4 (Dppa4) pAb (Rabbit, Ammonium Sulfate Purified) | |
| Cat No | CAC-TMD-PB-DP4 | |
| Description | DPPA4 (developmental pluripotency associated 4) is a highly expressed gene in early embryos and ES cells. DPPA4 binds to transcriptionally active chromatin and suppresses the differentiation of ES cells into primitive ectoderm (future endoderm). Source: Professor Hirofumi Teraoka, Department of Pathology and Biochemistry, Research Institute for Intractable Diseases, Tokyo Medical and Dental University. References: Masaki H, Nishida T, Kitajima S, Asahina K, and Teraoka H.: J. Biol. Chem. 282, 33034-33042 (2007). |
|
| Host | Rabbit | |
| Species specificity | MS | |
| Figure 1 | 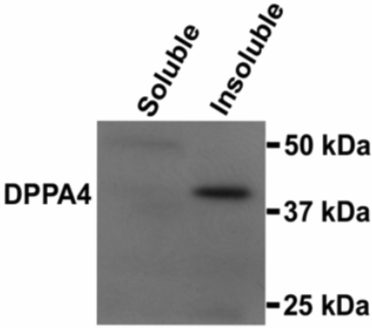 |
|
| Western blot analysis of DPPA4 in soluble and insoluble chromatin fractions. | ||
| Figure 2 | 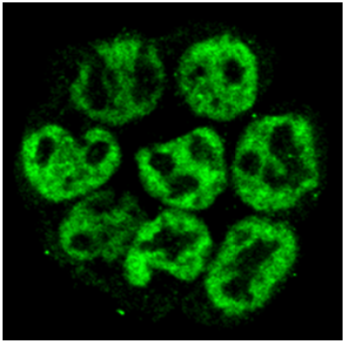 |
|
| Confocal image of ES cells immunostained with anti-DPPA4 antibody. | ||
| Product name | Anti Achaete-Scute Homolog 1 (ASH-1/MASH-1) pAb (Rabbit, Antiserum) | |
| Cat No | CAC-SK-T01-003 | |
| Description | Component of the RPD3C(L) histone deacetylase complex (HDAC). Responsible for the deacetylation of lysine residues on the N-terminal tails of core histones (H2A, H2B, H3 and H4). Histone deacetylation is a tag for epigenetic repression and plays an important role in transcriptional regulation, cell cycle progression and developmental events. ASH1 is necessary to repress HO in daughter cells to block mating-type switching through its binding to HO promoter 5'-YTGAT-3' sites. Also involved in pseudohyphal growth. References: Yoshida et al. (2001) Proprotein convertase PACE4 is down-regulated by the basic helix-loop-helix transcription factor hash-1 and MASH-1. Biochem. J. 360, 683-689. |
|
| Host | Rabbit | |
| Species specificity | HU MS | |
| Figure | 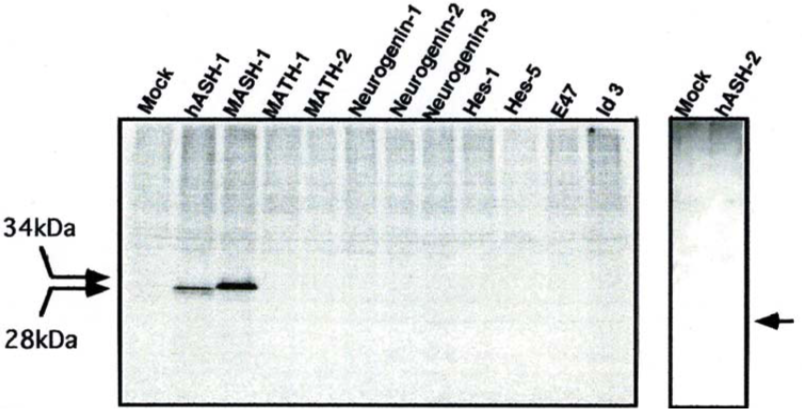 |
|
| - | ||
| Product name | Anti ASH-2 pAb | |
| Cat No | CAC-SK-T01-004 | |
| Description | Component of the Set-2/Ash-2 histone methyltransferase (HMT) complex. Required for di- and trimethylation at Lys-4 of histone H3, a mark associated with epigenetic transcriptional activation. Implicated in the epigenetic inheritance of lifespan over several generations. Functions as a transcriptional regulator. Acts in the germline to limit soma longevity, probably by regulating a lipid metabolism pathway that signals from the germline to the intestine, thereby preventing accumulation of mono-unsaturated fatty acids. References: Koide et al. The expression of proprotein convertase PACE4 is highly regulated by Hash-2 in placenta: Possible role for placenta-specific basic helix-loop-helix transcription factor, human achaete-scute homologue-2. J. Biochem. 134. 433-440. |
|
| Host | Rabbit | |
| Species specificity | HU MS | |
| Figure | 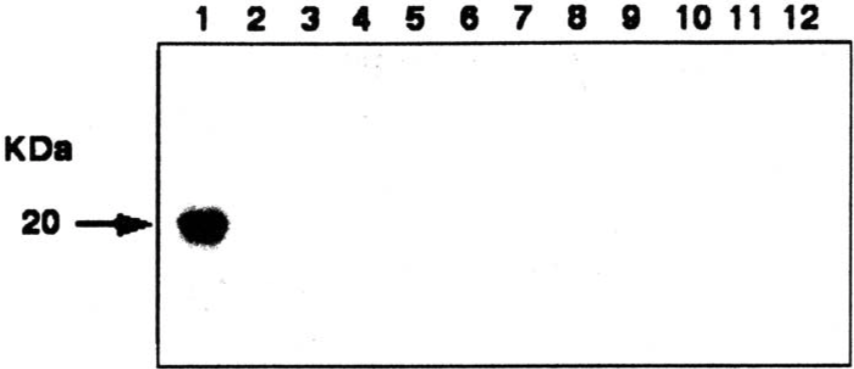 |
|
| Western blot. 1: hASH-2 2: hASH-1 3: MASH-1 4: MATH-1 5: MATH-2 6: neurogenin-1 7: neurogenin-2 8: neurogenin-3 9: Hes-1 10: Hes-5 11: E47 12: Id3 |
||
| Product name | Anti Transcription Activator BRG1 (SMARCA4) mAb (Clone 3G4) | |
| Cat No | CAC-CE-021A | |
| Description | Brm-related gene-1 (Brg1) is a catalytic subunit of the SWI/SNF chromatin remodeling enzyme complex that has ATPase activity. This complex facilitates chromatin remodeling for gene expression by utilizing energy from ATP hydrolysis. It is well known that the SWI/SNF chromatin remodeling enzyme complex is essential for cell differentiation, cell cycle regulation, and embryogenesis. References: 1) Ohkawa et al. (2007) J. Biol. Chem. 282:6564-6570. 2) Ohkawa et al. (2010) Hybridoma. 6:463-466. |
|
| Host | RT | |
| Species specificity | HU MS MKY | |
| Figure 1 | 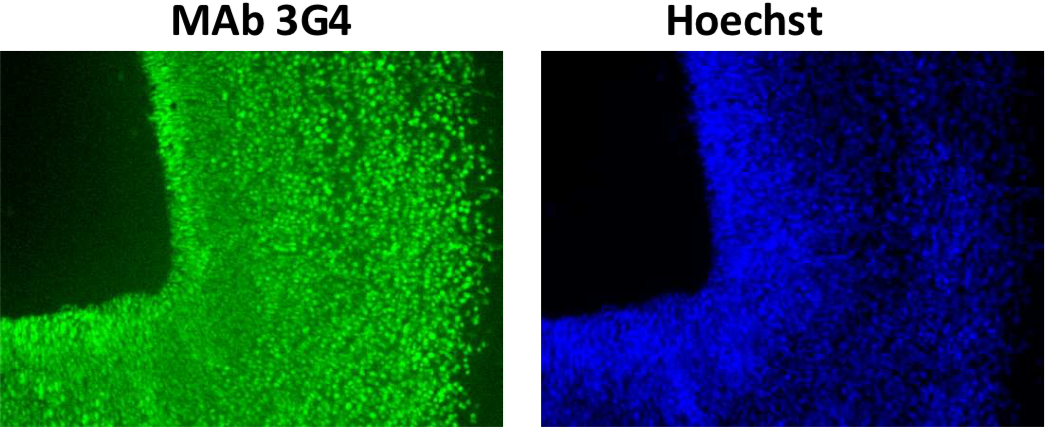 |
|
| Immunohistochemistry/immunofluorescence analysis of Brg1 antibody (3G4) on mouse E12.5 hind brain. | ||
| Figure 2 | 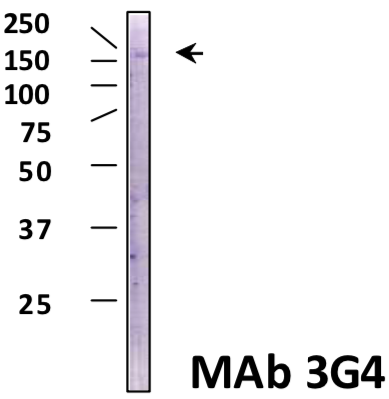 |
|
| Immunoblot analysis of Brg1 antibody (3G4) on HeLa cell extracts. | ||
| Product name | Anti Transcription Activator BRG1 (SMARCA4) mAb (Clone 5B7) | |
| Cat No | CAC-CE-021B | |
| Description | Brm-related gene-1 (Brg1) is a catalytic subunit of the SWI/SNF chromatin remodeling enzyme complex that has ATPase activity. This complex facilitates chromatin remodeling for gene expression by utilizing energy from ATP hydrolysis. It is well known that the SWI/SNF chromatin remodeling enzyme complex is essential for cell differentiation, cell cycle regulation, and embryogenesis. References: 1) Ohkawa et al. (2007) J. Biol. Chem. 282:6564-6570. 2) Ohkawa et al. (2010) Hybridoma. 6:463-466. This antibody is used in ref.1 and 2. |
|
| Host | RT | |
| Species specificity | HU MS MKY | |
| Figure 1 | 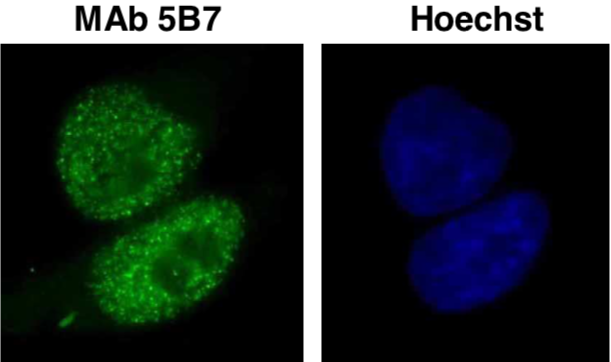 |
|
| Immunocytochemistry/Immunofluorescence analysis of Brg1 antibody (5B7) in HeLa cells. | ||
| Figure 2 | 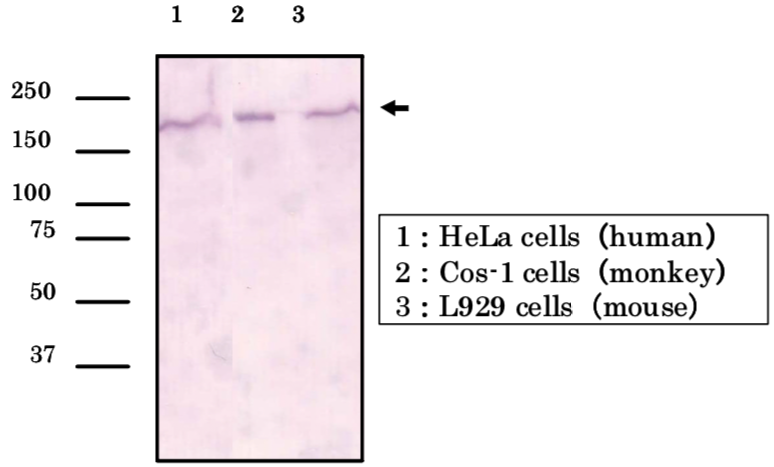 |
|
| Immunoblot analysis of Brg1 antibody (5B7) on cell extracts. |
||
| Product name | Anti SWI/SNF-related Matrix-associated Actin-dependent Regulator of Chromatin subfamily B member 1 (SMARCB1/INI1/BAF47) mAb (Clone 2C2) | |
| Cat No | CAC-CE-022A | |
| Description | SMARCB1/INI1/BAF47 is a core component of the BAF (SWI/SNF) complex. This ATP-dependent chromatin-remodeling complex plays important roles in cell proliferation and differentiation, in cellular antiviral activities and inhibition of tumor formation. The BAF complex is able to create a stable, altered form of chromatin that constrains fewer negative supercoils than normal. References: Hollmann and Hornick (2011) Am J Surg. |
|
| Host | RT | |
| Species specificity | HU MS | |
| Figure 1 | 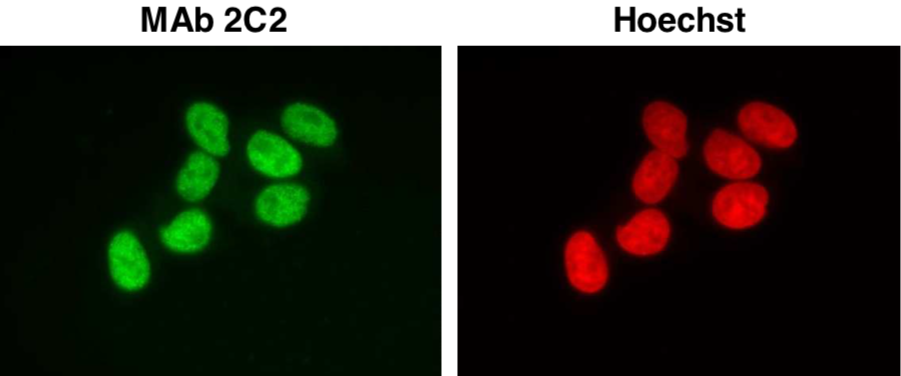 |
|
| Immunocytochemistry/Immunofluorescence analysis of Ini1/hSNF5/BAF47 antibody (2C2) in HeLa cells. | ||
| Figure 2 | 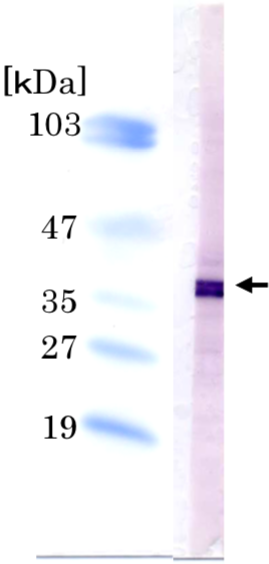 |
|
| Immunoblot analysis of Ini1/hSNF5/BAF47 antibody (2C2) on HeLa cell extracts. | ||
| Product name | Anti Condensin-2 Complex Subunit H2 (CAP-H2) mAb (Clone 5F2G4) | |
| Cat No | CAC-CE-024A | |
| Description | CAP-H2 is regulatory subunit of the condensin-2 complex, a complex that seems to provide chromosomes with an additional level of organization and rigidity and in establishing mitotic chromosome architecture. References: Wood et al. (2010) Nat. Rev. Genet. 11:391-404. |
|
| Host | RT | |
| Species specificity | HU MS | |
| Figure 1 | 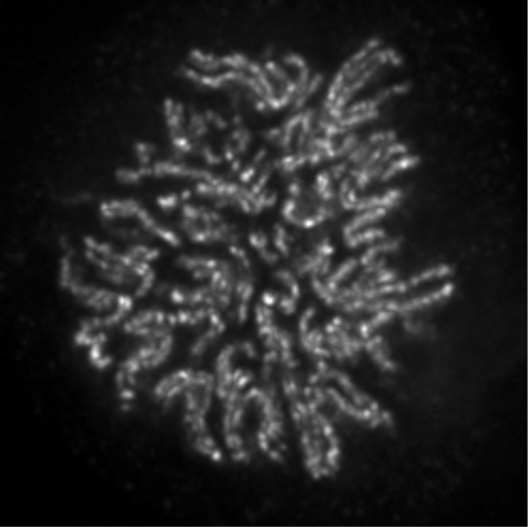 |
|
| Immunocytochemistry/Immunofluorescence analysis of condensin-2 complex subunit CAP-H2 antibody (5F2G4) on HeLa cells. | ||
| Figure 2 | 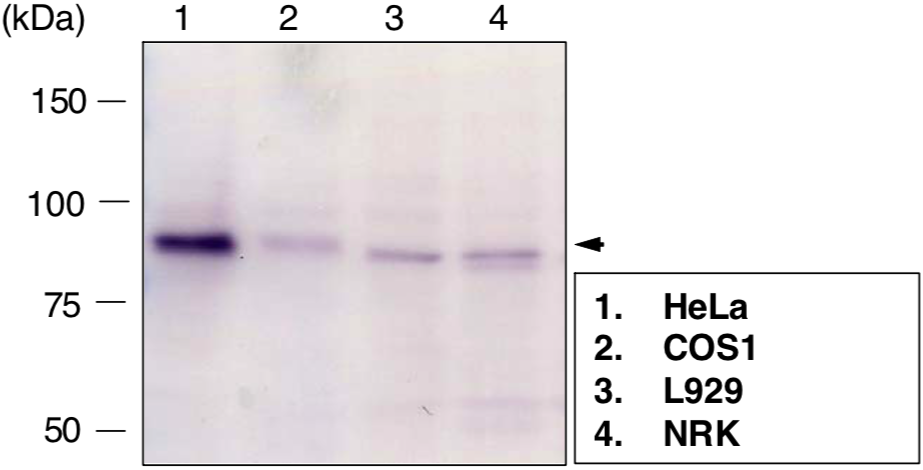 |
|
| Immunoblot analysis of condensing-2 complex subunit CAP-H2 antibody (5F2G4) on different cell extracts. | ||
| Product name | Anti Chromodomain-Helicase-DNA-Binding Protein 1 (CHD1) mAb (Clone 2F11H5) | |
| Cat No | CAC-CE-025A | |
| Description | CHD1 belongs to a subfamily of the CHD family. It possesses a chromodomain, a helicase domain, and a DNA-binding domain. The CHD family regulates gene expression by contributing to ATP-dependent chromatin remodeling. CHD1 occurs in transcriptionally active regions and alters chromatin structure. References: Yoshimura et al. (2010) Hybridoma (Larchmt). 29(3):237-40. PMID: 20568999 |
|
| Host | RT | |
| Species specificity | HU MS | |
| Figure 1 | 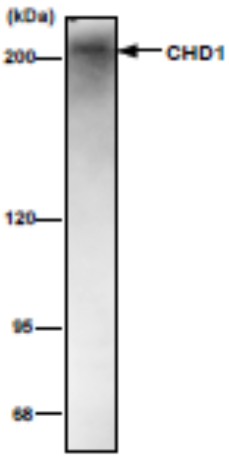 |
|
| Immunoblot analysis of NIH3T3 nuclear extract using CDH1 antibody (2F11H5). | ||
| Figure 2 | 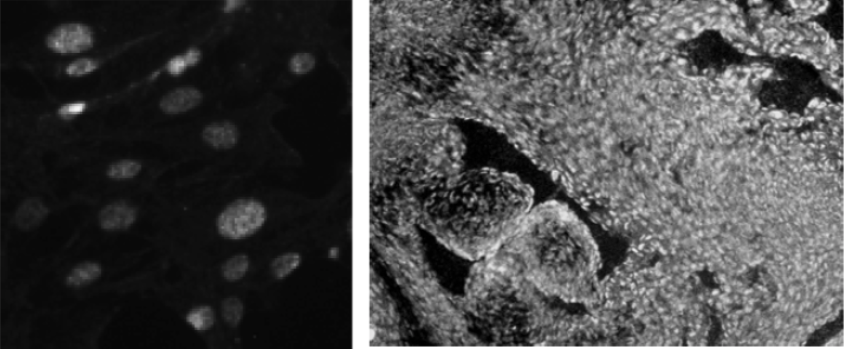 |
|
| Immunofluorescence analysis of NIH3T3 (left) and E12.5 mouse heart (right) using CHD1 antibody (2F11H5). | ||
| Product name | Anti Chromodomain-Helicase-DNA-Binding Protein 2 (CHD2) mAb (Clone 6D2) | |
| Cat No | CAC-CE-026A | |
| Description | CHD2 belongs to the CHD family which is characterized by chromodomains, helicase domains and DNA-binding domains. The CHD family is involved in gene expression and transcription by ATP-dependent chromatin remodeling. Analysis of CHD2 mutant mice revealed that CHD2 is involved in development as well as hematopoiesis. References: Harada A, et al. (2010) Hybridoma (Larchmt). 29(2):173-7. PMID: 20443711. |
|
| Host | RT | |
| Species specificity | HU MS | |
| Figure 1 | 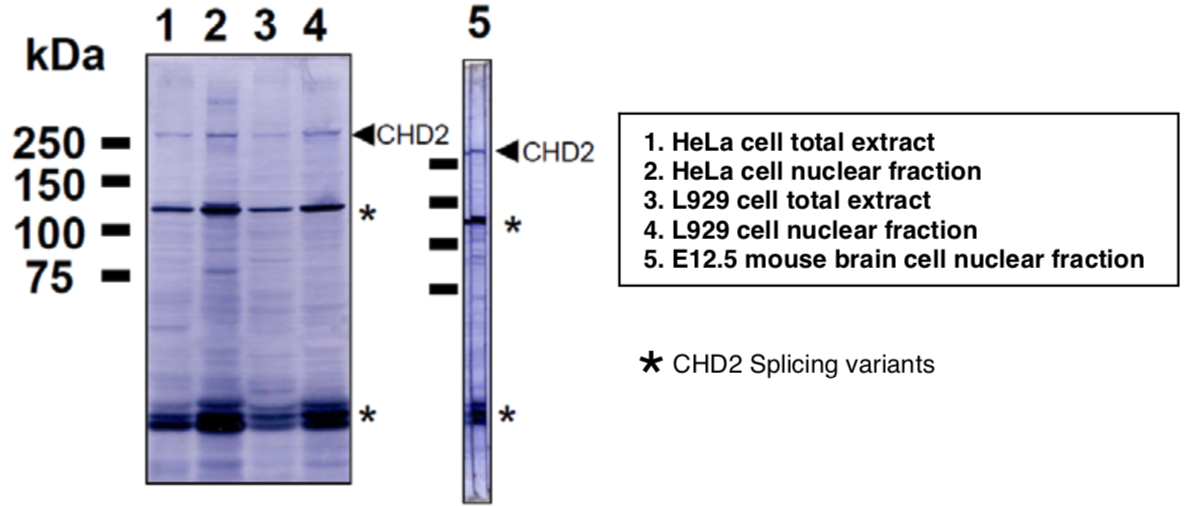 |
|
| Immunoblot analysis of Hela and L929 cell extracts and E12.5 mouse brain extracts using CDH2 antibody (6D2). | ||
| Figure 2 | 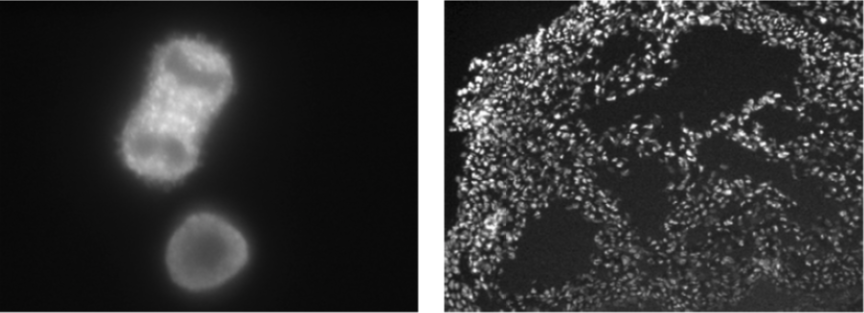 |
|
| Immunofluorescence analysis of HeLa cells (left) and E12.5 mouse heart (right) using CHD2 antibody (6D2). | ||
| Product name | Anti ATP-dependent RNA Helicase A (DHX9) mAb (Clone 8E3) | |
| Cat No | CAC-CE-028A | |
| Description | Dhx9/NDHII/RHA is a member of the DEAH family of proteins, which possess a double-stranded RNA-binding domain (dsRBD) and a helicase domain. The DEAH protein family plays a critical role in RNA metabolism. DEAH family members function as ATP-dependent RNA helicases and regulation of transcription. References: Kotani et al. (2010) Hybridoma 3:259-261. |
|
| Host | RT | |
| Species specificity | HU MS RT BOV MKY HAM | |
| Figure 1 | 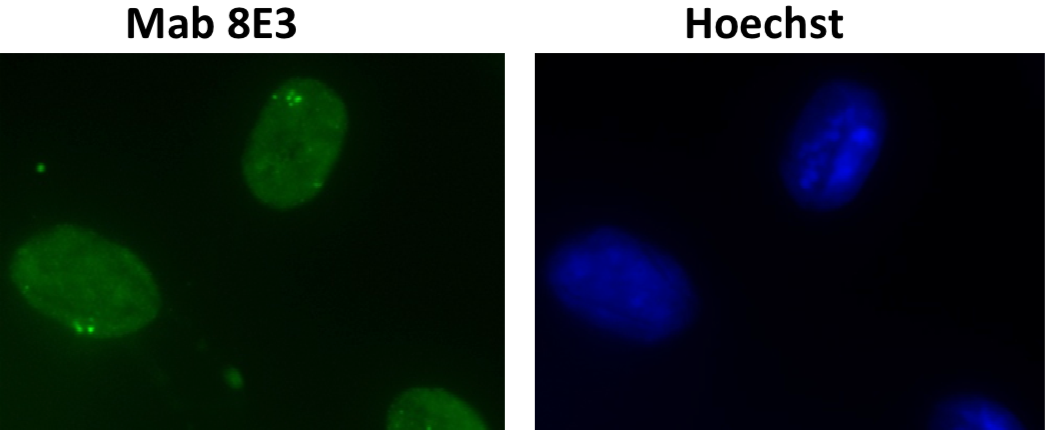 |
|
| Immunocytochemistry/Immunofluorescence analysis of DHX9 /RNA helicaseA antibody (8E3) on mouse L929 cells. | ||
| Figure 2 | 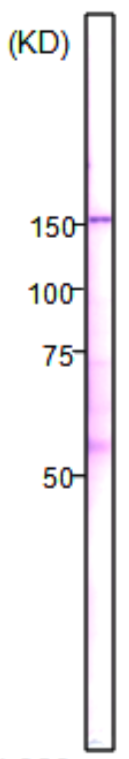 |
|
| Immunoblot analysis of DHX9 /RNA helicaseA antibody (8E3) on L929 cell nuclear extracts. | ||
| Product name | Anti DNA-Directed RNA Polymerase II Subunit RPB1 (POLR2A) CTD Phosopho-Ser2 mAb (Clone 3E7C7) | |
| Cat No | CAC-CE-030A | |
| Description | RNA polymerase II (RNAPII) transcribes all protein-coding genes and many non-coding genes. Its activity correlates with the phosphorylation state of RPB1, the large catalytic subunit of RNAPII. RPB1 has an unusual C-terminal domain (CTD) that consists of repeats of the heptapeptide consensus sequence N-Tyr-Ser-Pro-Thr-Ser-Pro-Ser-C, of which there are 52 copies in mammals. The amino acids in these repeats are potential targets for post-translational modification, such as phosphorylation and glycosylation. References: Odawara et al. (2011) BMC Genomics. 12:516. PMID: 22011111. |
|
| Host | RT | |
| Species specificity | HU MS RT MKY HAM | |
| Figure 1 |  |
|
| The composition of CTD peptides and their reactivity with RNA polymerase 2, CTD Ser2ph antibody 3E7C7. | ||
| Figure 2 | 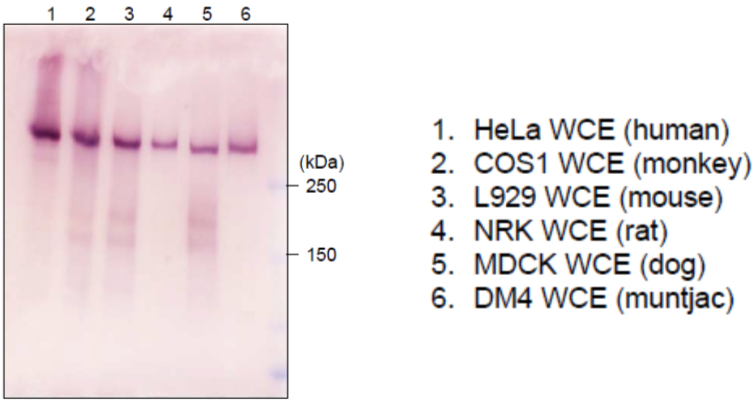 |
|
| Immunoblot analysis of mammalian cells extracts using RNA polymerase 2, CTD Ser2ph antibody 3E7C7. | ||
| Figure 3 | 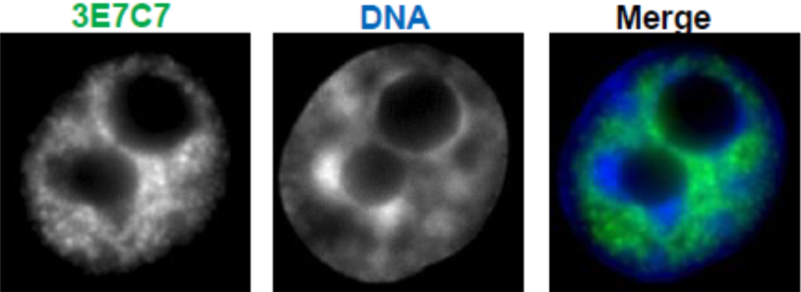 |
|
| Immunocytochemical analysis of HeLa cells using RNA polymerase 2, CTD Ser2ph antibody 3E7C7. | ||
| Product name | Anti DNA-Directed RNA Polymerase II Subunit RPB1 (POLR2A) CTD Phosopho-Ser5 mAb (Clone 1H4B6) | |
| Cat No | CAC-CE-031A | |
| Description | RNA polymerase II (RNAPII) transcribes all protein-coding genes and many non-coding genes. Its activity correlates with the phosphorylation state of RPB1, the large catalytic subunit of RNAPII. RPB1 has an unusual C-terminal domain (CTD) that consists of repeats of the heptapeptide consensus sequence N-Tyr-Ser-Pro-Thr-Ser-Pro-Ser-C, of which there are 52 copies in mammals. The amino acids in these repeats are potential targets for post-translational modification, such as phosphorylation and glycosylation. References: Odawara et al. (2011) BMC Genomics. 12:516. PMID: 22011111. |
|
| Host | RT | |
| Species specificity | HU MS RT MKY HAM | |
| Figure 1 | 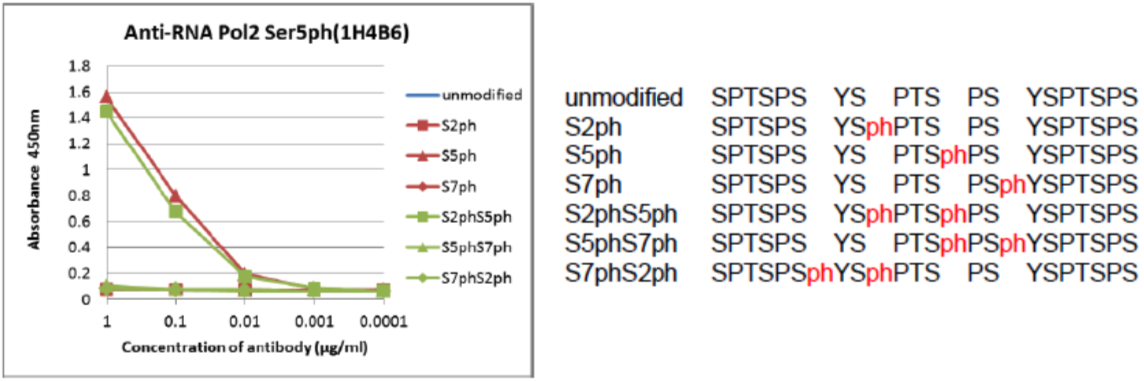 |
|
| The composition of the CTD peptides and the reactivity of RNA polymerase 2, CTD Ser5ph antibody 1H4B6. | ||
| Figure 2 | 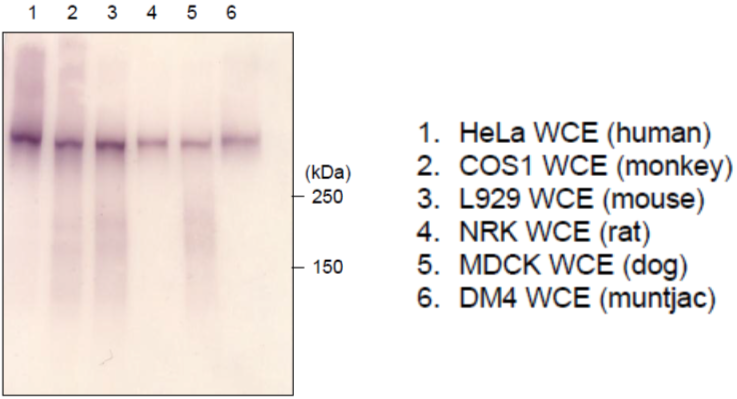 |
|
| Immunoblot analysis of mammalian cells extracts using RNA polymerase 2, CTD Ser5ph antibody 1H4B6. | ||
| Figure 3 | 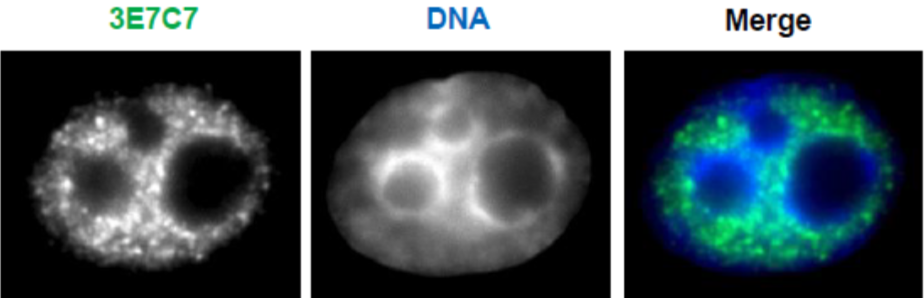 |
|
| Immunocytochemical analysis of HeLa cells using RNA polymerase 2, CTD Ser5ph antibody 1H4B6. | ||
| Drosophila chromatin |
||
| FACT (Facilitates Chromatin Transcription) is a protein complex involved in chromatin remodeling and comprises two subunits, SSRP1 and SPT16. A recent Drosophila study suggested that FACT forms a complex with a DNA binding protein, GAGA factor, to regulate sequence-specific changes in chromatin structure. SSRP1 and SPT16 have come to be noted as important new factors linking FACT and GAGA factors. | ||
| Product name | Anti Drosophila FACT Complex Subunit Ssrp1 (dSSRP1) pAb (Rabbit, Antiserum) | |
| Cat No | CAC-NIG-L1-SHA1 | |
| Description | dSSRP1 is the Drosophila ortholog of SSRP1. Studies have revealed that dSSRP1 facilitates various chromatin transactions such as transcriptional regulation, DNA replication and histone replacement. This antibody against dSSRP1 is a powerful tool for Western blots, immunostaining, immunoprecipitation and chromatin immunoprecipitation. Source: Dr. Susumu Hirose, National Institute of Genetics. References: 1) Shimojima, T. et al. , (2003) Genes & Dev . 17, 1605-1616. 2) Saunders, A. et al. , (2003) Science , 301, 1094-1096. 3) Nakayama, T. et al. , (2007) Genes & Dev . 21, 552-561. |
|
| Host | Rabbit | |
| Species specificity | Drosophila | |
| Figure 1 | 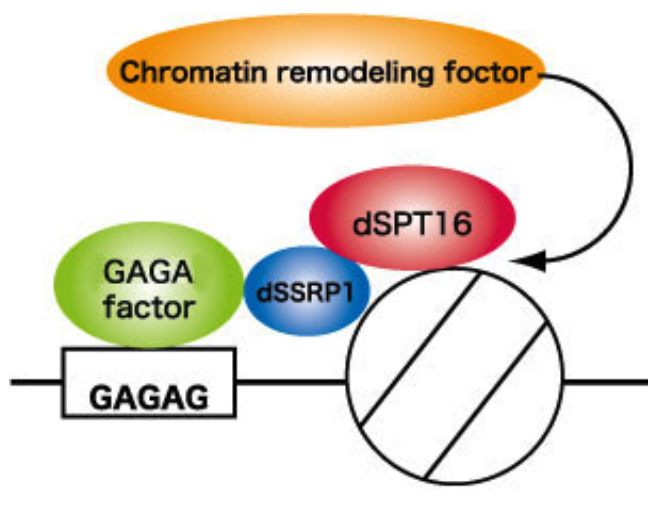 |
|
| Sequence-specific chromatin remodeling by GAGA factor-FACT complex. | ||
| Figure 2 | 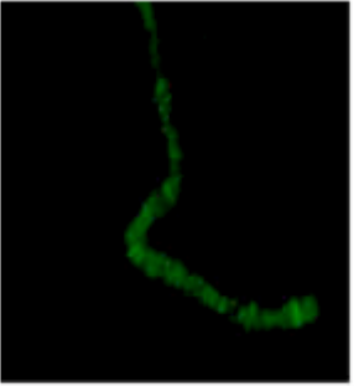 |
|
| Immunostaining of a polytene chromosome with anti dSSRP1 antibody. | ||
| Product name | Anti Drosophila FACT Complex Subunit Spt16 (dSPT16/DRE4) pAb (Rabbit, Antiserum) | |
| Cat No | CAC-NIG-L1-SHA2 | |
| Description | dSPT16 is the Drosophila ortholog of SPT16. Studies have revealed that dSPT16 facilitates various chromatin transactions such as transcriptional regulation, DNA replication and histone replacement. This antibody against dSPT16 is a powerful tool for Western blots, immunostaining (2), immunoprecipitation and chromatin immunoprecipitation. Source: Dr. Susumu Hirose, National Institute of Genetics. References: 1) Shimojima, T. et al. , (2003) Genes & Dev . 17, 1605-1616. 2) Saunders, A. et al. , (2003) Science , 301, 1094-1096. 3) Nakayama, T. et al. , (2007) Genes & Dev . 21, 552-561. |
|
| Host | Rabbit | |
| Species specificity | Drosophila | |
| Figure 1 | 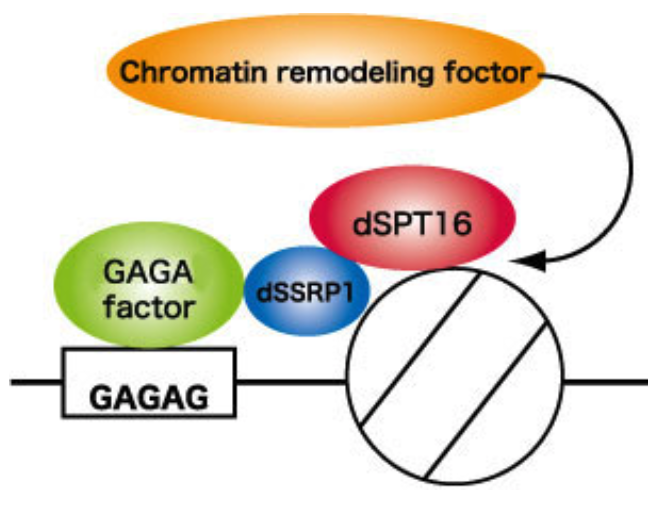 |
|
| Sequence-specific chromatin remodeling by GAGA factor-FACT complex. | ||
| Figure 2 | 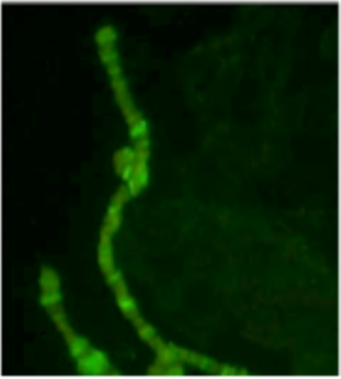 |
|
| Immunostaining of a polytene chromosome with anti dSPT16 antibody. | ||
